Actinomycetes as a Possible Source of Bioremediation of Heavy Metal Cadmium from Contaminated Soil
1
Department of Microbiology,
Atmiya University,
Rajkot,
Gujarat
India
Corresponding author Email: mousumi.das@atmiyauni.ac.in
DOI: http://dx.doi.org/10.12944/CWE.19.1.26
Copy the following to cite this article:
Musani T, Das M. Actinomycetes as a Possible Source of Bioremediation of Heavy Metal Cadmium from Contaminated Soil. Curr World Environ 2024;19(1). DOI:http://dx.doi.org/10.12944/CWE.19.1.26
Copy the following to cite this URL:
Musani T, Das M. Actinomycetes as a Possible Source of Bioremediation of Heavy Metal Cadmium from Contaminated Soil. Curr World Environ 2024;19(1).
Download article (pdf)
Citation Manager
Publish History
Select type of program for download
| Endnote EndNote format (Mac & Win) | |
| Reference Manager Ris format (Win only) | |
| Procite Ris format (Win only) | |
| Medlars Format | |
| RefWorks Format RefWorks format (Mac & Win) | |
| BibTex Format BibTex format (Mac & Win) |
Article Publishing History
| Received: | 2023-10-17 |
|---|---|
| Accepted: | 2024-01-30 |
| Reviewed by: | 
 Jaspal Kaur
Jaspal Kaur
|
| Second Review by: |

 Sandip Sharma
Sandip Sharma
|
| Final Approval by: | Dr. Gopal krishan |
Introduction
Over the recent past, the prompt escalation of heavy metals as contaminants poses a growing threat to the environment and through the accumulation in soil and subsequent uptake by plants, heavy metals are generating devastation in the environment, particularly in the agricultural sector1. This might be caused by the prolonged use of Phosphate fertilizers, the utilization of sewage sludge, industrial waste, and improper irrigation of agricultural regions2. Aluminum (Al), lead (Pb), cadmium (Cd), and silver (Ag) are examples of heavy metals that are poisonous to living things and have no biological use3. Majority Cd particles are absorbed by plant roots in accordance with portability, bioavailability, and concentration. Cd accumulates in the roots, shoots, and edible sections of plants after entering plant cells by a variety of carriers, including Calcium (Ca) channels4. Additionally, the buildup of Cd in plants has harmful consequences, such as the repression of some activities (mineral transport, photosynthetic apparatus, and nutrient intake). Additionally, it may prevent the entry of Fe into plant shoots. In recent decades, the crucial need for heavy metal pollution removal has expanded. Frequently utilized methodologies for the elimination of toxic metals from the surroundings encompass chemical precipitation, reverse osmosis, ion exchange, ultrafiltration, and electrodialysis. Unfortunately, they are frequently ineffective and quite pricey. They may also release poisonous chemicals, which are undesirable and unprofitable5. Therefore, there is a major need for the discovery of low-cost, exceedingly successful, and selective substitutes that can reduce heavy metal toxicity to levels that are appropriate to the environment. Thus, bioremediation is an appealing and effective cleansing method to get rid of toxic waste from a contaminated environment6. Environmental heavy metals are treated using bioremediation, which employs a variety of microbes containing bacteria, yeast, fungi, algae, and higher plants as its major treatment agents. The most applicable among these is bacteria as it is both inexpensive and environmentally beneficial. It may be done on-site, costs less to treat, has less impact on the environment, and does not result in secondary pollution7. The most significant groupings of microorganisms, the actinobacteria, are in charge of the bioremediation of hazardous heavy metals as well as the destruction and transformation of organic and metal substrates (Cd, Cr, Cu, Pb, Hg and Zn). Streptomycetes, a genus of actinobacteria, play a vital job in the extraction of metals from polluted water and soil. This capability is credited to their metabolic diversity, specific growth characteristics, spore-forming capacity, mycelia formation, and the comparatively quickly colonization of substrates. Because they produce a variety of metal ion chelators, like siderophores, that gives safety from the detrimental impact of heavy metals or selective absorption for particular metabolic activities, actinomycetes have the capacity to survive in environments polluted by metals8. To ascertain the indigenous bacterial strains' resistance to cadmium, the current study seeks to extract and identify them from heavy metal-contaminated soil. The traits of the strains' physiology and biochemistry were used to define them.
Materials and methods
Collection and analysis of soil sample
The soil specimen was obtained from agricultural site in Jamnagar district, Gujarat with a latitude and longitude of the area, 22°18’11” N, 70°18’57” E. Soil sample was taken from the surface to a depth of 13-15 cm. Soil samples were gathered using zippered bags for the sampling process. Stone fragments and plant remains were manually removed and cleaned. A 2 mm brass sieve was used to filter soil sample and air dried. Soil analysis was performed at Envitro Soil Laboratory, Rajkot, which includes soil pH, Electrical Conductivity, Total Organic Carbon, Nitrogen, Phosphorous, Potash, Cation Exchange Capacity, Moisture and heavy metals such as Zinc (Zn), Cadmium (Cd) and Copper (Cu).
Enrichment of soil sample
By using a sterile spatula, sample was deposited in sterile Ziplock bags. Soil sample was kept at 4° C and brought to the lab for processing right away. For the enrichment of actinomycetes, the sample was treated to a chemical treatment with CaCO3 9.
Isolation of actinomycetes
One gram of soil was mixed with 9 ml of autoclaved N-saline, which repeatedly diluted to dilutions of 10-3, 10-4, and 10-5. On Starch-Casein Agar (SC) medium, 0.1 mL of each dilution were dispersed. Petri dishes were kept in a chamber for culturing at 28°C for 7–12 days after sub-culturing isolated actinobacteria10.
Primary Screening of metal tolerant actinomycetes by plate assay method
By adding 100 mg/L of CdCl2 to sterilized SC medium, the isolated actinomycetes colonies were tested for heavy metal resistance to cadmium. The isolated actinomycetes colonies were subjected to a purification process and cultured at 28° C for 10-12 days on SC agar medium. The greatest concentration of heavy metal that allows for development after maximum concentration (7-12 days) that may be tolerated. Isolated actinomycetes colonies that had grown on SC agar supplemented with cadmium were then subjected to increasing levels of cadmium (100, 500, 1000, and 1500 mg/L). When complete inhibition of growth was observed on SC agar with metal supplementation, the testing for microorganism tended tolerance came to an end11.
Secondary Screening of metal tolerant actinomycetes by agar well diffusion method
Test was conducted in SC agar medium plates treated with CdCl2 at concentrations of 100, 500, 1000 and 1500 mg/L. In this instance, wells were created by aseptically removing strips of agar from the center of Petri dishes and then filling them with 500 µL of 100, 500, 1000 and 1500 mg/L CdCl2 solutions. Isolates were inoculated by streaking perpendicularly to the wells. Petri dishes were cultured at 28°C for 10-12 days. The ratio of metal tolerance exhibited by the actinomycetes strain was estimated by determining the proportion in between the length of the observed growth (in millimeters) and the total length of the inoculated streak.
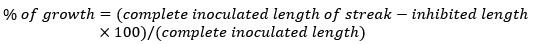
Consequently, the inhibition exerted by the metal heightened with the greater distance of the growing colony from the well's edge. The proximity of microbial growth to the well served as the qualitative measure of metal resistance.12.
Spore Optimization
Spore optimization was carried out by adding a drop of tween 80 reagent mixed with 5 ml distilled water on a slant. The solution was mixed homogenously and allowed it to settle. 1 ml of supernatant was taken and serially diluted to dilution of 10-5 on SCA media supplemented with Cd and without Cd. By using a spread plate approach, the viability of the spores was assessed. After the petri dishes were cultured for 11-13 days at 28°C, colony forming units (CFUs) were counted. Spore counts were performed and compared with control13. High spore forming actinomycetes strain was selected for molecular identification. Isolates were proceeded further for their spore counting by using this formula,

Morphological and Biochemical characterization of actinomycetes isolates
The morphological and biochemical properties like colony characteristics, Gram staining, Catalase activity, Carbon utilization property, Citrate utilization activity, Starch hydrolysis, Gelatine hydrolysis, Casein hydrolysis, Methyl Red, Voges-Proskauer, Nitrate reduction of strain AMT14, AMT35, AMT37 and AMT52 were performed14.
Molecular identification of actinomycetes isolates
The genomic material of highly spore-forming actinomycetes was isolated, and its DNA content was quantified using a 1.0% Agarose Gel, revealing one band of large-molecular-size DNA. A targeted gene segment was then amplified through PCR, yielding a distinct amplicon band upon electrophoresis. Subsequent purification using a column method removed impurities from the PCR product. DNA sequencing of the purified amplicon was performed applying the BDT v3.1 Cycle Sequencing Kit on an ABI 3500xl Genetic Analyzer having primer27 F. The resulting gene sequence underwent BLAST analysis against the NCBI GenBank database. The top ten sequences, based on maximum identity scores, were selected and matched using several alignment programs. Gene sequences were further compared with the GenBank database through the NCBI and BLAST platform. The sequencing methods were implemented by Gene Explore Diagnostics and Research Centre Private Limited in Ahmedabad, Gujarat15.
Results
Analysis of soil sample
The soil used in this study was contaminated by pesticide. Total Zinc (Zn) and Copper (Cu) contents demonstrated that it was not contaminated by these metals. On the other hand, it was contaminated by heavy metal Cadmium (Cd). Lower availability of metals in the soil occurs from low pH, which increases the mobility of metals in the soil. Nearly all biological and chemical processes that regulate terrestrial ecosystems are influenced by soil pH. The contents of soil physical characteristics and heavy metals in the investigation area are shown in Table 1 and 2.
Table 1: Physical and Chemical parameters of soil sample
Sr. No. | Tests | Result |
1 | pH | 6.26 |
2 | Electric Conductivity | 6.14 mS/Cm2 |
3 | Total Organic Carbon | 17.75 % |
4 | Nitrogen | 0.23 % |
5 | Phosphorus | 0.002 % |
6 | Potash | 0.02 % |
7 | Moisture | 25.97 % |
8 | Cation Exchange Capacity | 17.88 meq |
Table 2: Concentration of Heavy metals in soil sample
Sr. No. | Tests | Results (mg/kg) | WHO limits (mg/kg) |
1 | Zinc (Zn) | 7.88 | 50 |
2 | Cadmium (Cd) | 14 | 0.8 |
3 | Copper (Cu) | 0.23 | 36 |
The physicochemical analysis of samples contaminated with pesticides has unveiled a substantial concentration of the heavy metal cadmium in the studied site. Moreover, the acidic pH and elevated salinity observed are intricately linked to the utilization of pesticides.
Isolation of Actinobacteria and Screening of Metal Resistant Strains
A sample of soil taken from an agricultural site yielded fifty-three actinobacterial colonies in total. Four actinomycetes strains were resistant to the heavy metal Cadmium out of the total number of actinomycetes strains that were isolated and cultured in starch casein agar with metal supplements. All the four bacterial colonies that were Cadmium resistant were tolerant up to 1000 mg/L, However, at elevated concentrations of metal salts, the growth of these isolates was impeded. None of them were tolerant up to 1500 mg/L of cadmium.
Secondary screening of actinobacteria by agar well diffusion method
Each individual isolate exhibited a distinct tolerance level compared to others when exposed to the same metal. Out of fifty-three isolates AMT14, AMT35, AMT37 and AMT52 were resistant at 1000mg/L. The total length of the inoculated streak measured 37 mm. The tolerance of cadmium among various actinomycetes isolates was determined based on the length of the growing streak. The growth percentage was computed using the specified formula and was later depicted graphically.
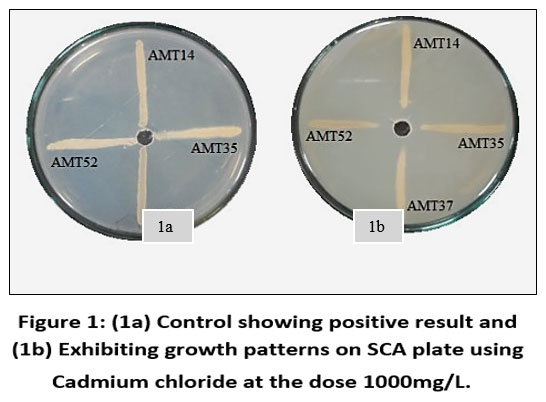 | Figure 1: (1a) Control showing positive result and (1b) Exhibiting growth patterns on SCA plate using Cadmium chloride at the dose 1000mg/L.
|
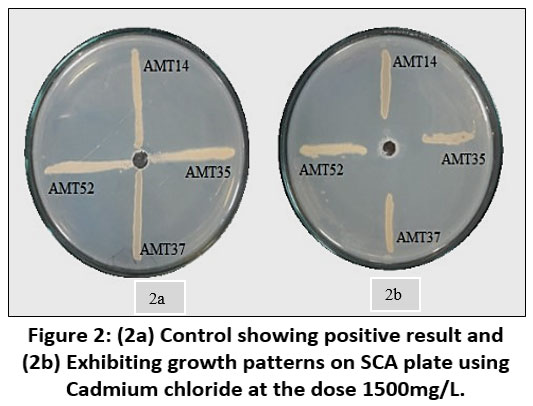 | Figure 2: (2a) Control showing positive result and (2b) Exhibiting growth patterns on SCA plate using Cadmium chloride at the dose 1500mg/L.
|
Table 3: % of isolates' development on SCA plates.
Sr. No. | Actinomycetes colonies | Assess the growth % on agar plates exposed to varying concentrations of CdCl2 | |
| 1000 ppm | 1500 ppm | |
1. | AMT14 | 84.76 | 54.05 |
2. | AMT35 | 80.13 | 53.02 |
3. | AMT37 | 87.24 | 86.28 |
4. | AMT52 | 78.16 | 63.92 |
Note: AMT= Strain number, CdCl2= Cadmium Chloride
 | Figure 3: Graphical presentation of % of growth by all strains at different metal level.
|
Spore Optimization
All four actinomycetes isolates were able to grow on SC agar media and able to produce spores. AMT52 having highest spore productivity when compared to control. Thus, it was selected as potent heavy metal tolerant isolates.
Table 4: Highest spore productivity by all bacterial strains
Sr. No. | Name of strain | Dilution
| Day of optimization | Control (spores/ml) | Heavy Metal (spores/ml) |
1. | AMT14 | 10-5 | 6th | 28 × 105 | 21 × 105 |
2. | AMT35 | 10-5 | 5th | 46 × 105 | 8 × 105 |
3. | AMT37 | 10-5 | 6th | 15 × 106 | 8 × 106 |
4. | AMT52 | 10-5 | 5th | 39 × 105 | 23 × 105 |
Note: AMT= Strain number
Identification of Metal Tolerance
Actinomycetes
From soil sample, the isolated actinomycetes were recovered. The morphological traits are still often employed to identify taxa. Microscopic examination of gram-stained slides revealed gram-positive actinomycetes with purple color, rod shape and filamentous structure. Biochemical tests were used to identify the type of bacteria present in isolates.
 | Table 5: Results of Actinomycetes colonies on SCA media and its Gram’s staining view under microscope
|
Note: AMT= Strain number
Table 6: Colony characteristics of potent actinomycetes
Strain No. | +/- | Shape | Size | Color | Texture | Elevation | Optical Properties |
AMT14 | +ve | Circular | Small | Chalky White | Rough | Slight Raised | Opaque |
AMT35 | +ve | Circular | Small | Chalky White | Rough | Slight Raised | Opaque |
AMT37 | +ve | Circular | Medium | Gray | Smooth | Slight Raised | Opaque |
AMT52 | +ve | Circular | Small | Gray | Smooth | Slight Raised | Opaque |
Note: AMT=Strain number, +ve= Gram positive bacteria and -ve= Gram negative bacteria
Table 7: Results of biochemical tests of potent Actinomycetes
Sr. No. | Tests | AMT14 | AMT35 | AMT37 | AMT52 |
1. | Catalase | + | + | + | + |
2. | Citrate utilization | + | - | + | + |
3. | Starch hydrolysis | + | + | + | + |
4. | Gelatine hydrolysis | + | - | - | - |
5. | Casein hydrolysis | - | - | - | - |
6. | Methyl Red | + | - | + | - |
7. | Voges Proskauer | + | - | - | + |
8. | Nitrate reduction | + | + | + | + |
Note: +=Positive and -= Negative
Table 8: Results of biochemical tests of potent Actinomycetes
Sr. No. | Sugars | AMT14 | AMT35 | AMT37 | AMT52 |
1. | Glucose | + | + | + | + |
2. | Lactose | + | + | - | + |
3. | Maltose | + | + | + | + |
4. | Sucrose | + | + | + | + |
5. | Mannitol | - | + | - | - |
6. | Galactose | + | + | + | + |
7. | Fructose | + | + | + | + |
Note: +=Positive and -= Negative
AMT52 was selected for more detail’s studies as it was high spore forming bacterial strain and recognized as Streptomyces pactum, a species from the genus Streptomyces.
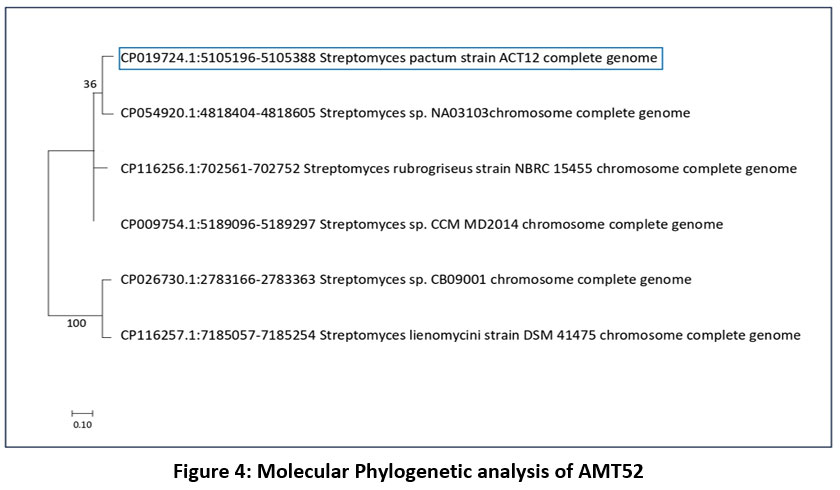 | Figure 4: Molecular Phylogenetic analysis of AMT52
|
Discussion
The usage of pesticides in agricultural regions has sharply increased in the past several years. Pesticides with composite and triple super phosphate had the highest levels of Cd16. Cadmium concentration in the studied region was highest than Zinc and Copper. After the enhancement of a soil sample, fifty-three actinomycetes that were tolerant of heavy metals were isolated; only four of the isolates expressed this sensitivity. The actinomycetes were selected in the presence of cadmium at concentrations of up to 1000 mg/L. All the isolates were recognized as Streptomyces spp. based on their morphological and biochemical characteristics. Strain AMT52 shown highest spore optimization in a presence of heavy metal cadmium compared to control, which was identified by molecular analysis as Streptomyces pactum. Bagot and others isolated actinomycetes from soil contaminated with cadmium and found best growth capacity of actinomycetes in a presence of 10 mg/L cadmium17. Hamedi and his colleagues highly identified resistance strains of actinomycetes genera showed resistance to 9.2mM CdCl2 including Streptomyces, Nonomuraea, Saccharothrix, Streptosporangium and Promicromonospora18. Rajivgandhi and others revealed Nocardiopsis dassonvillei as excellent biological source for heavy metal depletion from waste as they reduced heavy metal cadmium contamination up to 85.20%19. These studies suggest variability in the potency of bacteria towards heavy metals to which they are resistant. Actinobacterial spore cells produced good metal accumulating results in the current investigation, particularly for cadmium. To improve the bioremediation capacity of these strains, more research should be developed at the molecular level to assess and discover the mechanisms involved in the relationship between metal resistances and accumulation.
Conclusions
Areas contaminated with pesticides are deemed appropriate for isolating microorganisms resistant to heavy metals, serving as valuable reservoirs for potential bioaccumulation processes due to their high contamination levels with metallic elements. The process of heavy metal accumulation can be defined as the gathering of these components within the ecological systems. The restoration of such environments is notably intriguing, particularly through the application of eco-friendly remediation techniques aimed at enhancing their overall quality. We believe that several of the actinobacteria especially Streptomyces pactum we have chosen can contribute significantly to the clean-up of contaminated sites. However, additional investigation is required to improve our strains' biological process capabilities and to maximize their ability to eliminate heavy metals from polluted environments.
Acknowledgement
The authors express gratitude to the Head of the Department of Microbiology, Atmiya University, Rajkot, for valuable suggestions, infrastructure, and facilities provided. We thank Gene Explore Diagnostics and Research Centre Private Limited, Ahmedabad, for helping us with molecular identification of our potent strain. We also thank Envitro Soil Laboratory, Rajkot, for helping us with detailed soil investigation.
Conflict of Interest
The authors affirm that there are no conflicts of interest pertaining to the research presented in this paper, and they have no financial or personal relationships that could influence the work.
Funding Sources
The authors extend gratitude to the Government of Gujarat for the financial support received under its 'SHODH SCHEME' with Reference No. 202101241
Authors’ Contribution
In the manuscript the first and second author (corresponding author) has contributed in following sectors:
Conception or design of the work- The core idea behind the concept of work and incorporation of the components for the smooth progression of the research work is mentored by Dr.Mousumi Das and performance of the laboratory activity with documentation has completed by Ms.Tasnim Musani
Data collection- This part of the manuscript was completed by the research worker during her part of work in Ph.D. Idea of compilation and drafting was mentored by Dr. Mousumi Das
Data analysis and interpretation – This part was contributed by both the authors covering types of representation (selection of Table or graph); selection of images; documentation of the summarized data etc
Drafting the article–The drafting of the manuscript was performed by Ms.Tasnim Musani
Critical revision of the article- The analysis and addressing the comments and analysing the details of the manuscript components was guided by Dr. Mousumi Das and implemented by Ms. Tasnim Musani
Final approval of the version to be published–This part was mutually decided by First and corresponding author with sharing majority of the rights by Corresponding author
Data Availability Statement
The publication includes all datasets analyzed during the course of this research.
Ethics Approval Statement
This particular research study did not involve any animal or human experimentation.
References
- Alengebawy A, Abdelkhalek ST, Qureshi SR, Wang M-Q. Heavy metals and pesticides toxicity in agricultural soil and plants: Ecological risks and human health implications. Toxics. 2021;9(3):42.
CrossRef - Aladesanmi OT, Oroboade JG, Osisiogu CP, Osewole AO. Bioaccumulation factor of selected heavy metals in Zea mays. J Heal Pollut. 2019;9(24):191207.
CrossRef - Tumolo M, Ancona V, De Paola D, et al. Chromium pollution in European water, sources, health risk, and remediation strategies: An overview. Int J Environ Res Public Health. 2020;17(15):5438.
CrossRef - Zhou C, Ge N, Guo J, et al. Enterobacter asburiae reduces cadmium toxicity in maize plants by repressing iron uptake-associated pathways. J Agric Food Chem. 2019;67(36):10126-10136.
CrossRef - Ezeonuegbu BA, Machido DA, Whong CMZ, et al. Agricultural waste of sugarcane bagasse as efficient adsorbent for lead and nickel removal from untreated wastewater: Biosorption, equilibrium isotherms, kinetics and desorption studies. Biotechnol Reports. 2021;30:e00614.
CrossRef - Sharma, I. We are IntechOpen , the world ’ s leading publisher of Open Access books Built by scientists , for scientists TOP 1 % Bioremediation Techniques for Polluted Environment?: Concept. 2020.
- Jin, T., Shi, C., Wang, P., Liu, J., & Zhan, L. A review of bioremediation techniques for heavy metals pollution in soil. IOP Conference Series: Earth and Environmental Science. 2021;687(1).
CrossRef - Jagannathan S V, Manemann EM, Rowe SE, Callender MC, Soto W. Marine actinomycetes, new sources of biotechnological products. Mar Drugs. 2021;19(7):365.
CrossRef - Alferova I V, Terekhova LP. Use of the method of enriching of soil samples with calcium carbonate for isolation of Actinomyces. Antibiot i khimioterapiia= Antibiot chemoterapy [sic]. 1988;33(12):888-890.
- Anwar, S., Ali, B., & Sajid, I. Screening of rhizospheric actinomycetes for various in-vitro and in-vivo plant growth promoting (PGP) traits and for agroactive compounds. Frontiers in Microbiology. 2016;7:1–11.
CrossRef - De Guzman, M, L., Arcega, K, S., Cabigao, N, R., & Sia Su, G, L. Isolation and identification of heavy metal-tolerant bacteria from an industrial site as a possible source for bioremediation of cadmium, lead, and nickel. Advances in Environmental Biology. 2016;10(1):10–15.
- Albarracín, V. H., Amoroso, M. J., & Abate, C. M. Supplement 1. Chemie Der Erde. 2005; 65(1):145–156.
CrossRef - Queener SW, Capone JJ. Simple method for preparation of homogeneous spore suspensions useful in industrial strain selection. Appl Microbiol. 1974;28(3):498-500.
CrossRef - Erikson D. The morphology, cytology, and taxonomy of the actinomycetes. Annu Rev Microbiol. 1949;3(1):23-54.c trees. Mol Biol Evol. 1987;4(4):406-425.obiol. 1949;3(1):23-54.
CrossRef - Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4(4):406-425.
- Atafar Z, Mesdaghinia A, Nouri J, et al. Effect of fertilizer application on soil heavy metal concentration. Environ Monit Assess. 2010;160:83-89.
CrossRef - Bagot, D., Lebeau, T., Jezequel, K., & Fabre, B. Selection of microorganisms for bioremediation of agricultural soils contaminated by cadmium. Environmental Chemistry: Green Chemistry and Pollutants in Ecosystems. 2005;215–222.
CrossRef - Hamedi, J., Dehhaghi, M., & Mohammdipanah, F. (2015). Isolation of extremely heavy metal resistant strains of rare actinomycetes from high metal content soils in Iran. International Journal of Environmental Research. 2015;9(2):475–480.
- Rajivgandhi, G., Gnanamangai, B. M., Ramachandran, G., Chackaravarthy, G., Chelliah, C. K., Maruthupandy, M., Alharbi, N. S., Kadaikunnan, S., & Li, W. J. Effective removal of heavy metals in industrial wastewater with novel bioactive catalyst enabling hybrid approach. Environmental Research. 2022;204.
CrossRef






