Biodegradation of Low-and High-Density Polyethylene Films by Microbacterium barkeri SH20
Corresponding author Email: sharma.sakshifeb@gmail.com
DOI: http://dx.doi.org/10.12944/CWE.17.1.22
Copy the following to cite this article:
Sharma S, Mathur N, Singh A, Agarwal M. Biodegradation of Low-and High-Density Polyethylene Films by Microbacterium Barkeri SH20. Curr World Environ 2022;17(1). DOI:http://dx.doi.org/10.12944/CWE.17.1.22
Copy the following to cite this URL:
Sharma S, Mathur N, Singh A, Agarwal M. Biodegradation of Low-and High-Density Polyethylene Films by Microbacterium Barkeri SH20. Curr World Environ 2022;17(1).
Download article (pdf)
Citation Manager
Publish History
Select type of program for download
| Endnote EndNote format (Mac & Win) | |
| Reference Manager Ris format (Win only) | |
| Procite Ris format (Win only) | |
| Medlars Format | |
| RefWorks Format RefWorks format (Mac & Win) | |
| BibTex Format BibTex format (Mac & Win) |
Article Publishing History
| Received: | 28-10-2020 |
|---|---|
| Accepted: | 29-04-2022 |
| Reviewed by: | 
 Rizafizah Othaman
Rizafizah Othaman
|
| Second Review by: |

 Diego Fernando Ramirez Guerrero
Diego Fernando Ramirez Guerrero
|
| Final Approval by: | Dr. V P Tewari |
Introduction
The degradation of synthetic polymers like plastics is a major challenge due to its recalcitrant properties. Polyethylene (PE), which is a type of plastic, has become part of the humanity due to its excellent mechanical properties, oxygen & moisture barrier and low cost. It has been used for the manufacturing of carry bags, packaging materials, etc. (mainly in the form of thin films), and approximately 64% of all produced plastics is used to make polyethylene1, 2. Consequently, the occurrence of polyethylene in plastic waste is also exceedingly high. For instance, A revealed that approximately 6,300 million metric tons (Mt) of plastic waste is generated worldwide, out of which 9% is recycled, approximately 79% is stockpiled in landfills or is disposed in the surroundings and 12% is reduced to ashes3. The percentage of plastic waste generated with polypropylene, polyvinyl chloride, polystyrene, polyethylene terephthalate and other types are 18.5%, 10.7%, 12.3%, 8.5%, and 9.7%, respectively. Moreover, the highest component of plastic waste is generated from polyethylene with a 23% from low-density polyethylene (LDPE) & 17.3% from high-density polyethylene (HDPE)4. Growing levels of polyethylene waste, declining of landfill’s extent and very gradual degradation of polyethylene in environment are the main reasons for accumulation of plastic waste. Also, polyethylene degradation can last for long time frames. Furthermore, if polyethylene burnt carelessly, it releases severe toxic gases such as carbon monoxide, sulphur dioxide, phosgene, chlorine, and fatal dioxins5. For degradation of polyethylene waste there are few major types of degradation methods reported, such as, thermo-oxidative degradation, photo-degradation and biodegradation6. The abiotic methods such as thermo-oxidative and photo-degradation methods are likely to form toxic end products. Therefore, there is a need to find alternatives more benign to the environment for treating this type of plastic. In fact, the biological approach is currently proved to be a safe, effective and eco-friendly approach for plastic waste management7.
Biodegradation is a natural process where polymers can be used by microorganisms as carbon and energy sources, and it can prevent the formation of toxic end product8. The biological activities of microorganisms have been effectively used for stimulation of degradation in previous studies. Some microorganisms have specific abilities to colonize and form biofilms on the surface of the polymers which can modify, consume and change its properties that ultimately leads to degradation9.
Chemically, plastics are long chains of hydrocarbons possessing high molecular weight. Previous reports established that microorganisms isolated from motor-oil contaminated soil can degrade crude oil spill and many hydrocarbons compounds10, 11. As these motor-oil spill areas are contaminated with hydrocarbons thus, polyethylene degrading bacteria were expected to be present in these areas. Therefore, the aims of this study were to isolate and identify potential bacterial strain from motor-oil contaminated soil which was not isolated in previous researches for biodegradation of polyethylene as far. Also, there are many studies on biodegradation of either LDPE or HDPE but this study attempts to check bacterial isolate’s biodegradation efficiency on both LDPE and HDPE films.
Experimental
Oil-contaminated soil sampling
Soil samples for the isolation of polyethylene-degrading bacteria were collected from motor-oil contaminated soils near auto mechanical workshops in Transport Nagar, Jaipur, Rajasthan (India).
Isolation of Polyethylene Degrading Microorganisms
The following protocols were adapted12 for enrichment of polyethylene-utilizing microorganisms. The formulation of the enrichment medium per 1000ml were [Yeast Extract] - .06g, [(NH4)2 SO4] - 4g, [K2HPO4] - 2g, [KH2PO4] - 1g, [MgSO4.7H2O] - .5g. The pH modulated upto 7. Suspensions of soil samples were inoculated in the enrichment medium (100ml) with polyethylene powder-332119 (Sigma-Aldrich), at 1mg/ml, and then it was incubated in the rotary shaker for seven days at 32ºC with negative control. Flasks showing positive growth were subcultured consecutively from the original culture into fresh enrichment medium containing polyethylene powder as a carbon source.
For isolation of polyethylene degrading strains, emulsified basal medium agar plates with hexadecane (1ml/L) containing 0.1 gram of detergent were spread with subcultures selected from the enrichment medium and incubated for a week at 32ºC. Microbial colonies produce clear zones on medium plates supplemented by emulsified hexadecane were selected and streaked on fresh medium plates. Formulation of basal medium per 1000ml was: (NH4)2SO4 - 1g, Hexadecane - 1ml (M.Wt: 226.44g/mol) (Sigma-Aldrich), yeast extract - .06, K2HPO4 - 2.34g, MgSO4.7H2O - .2g, KH2PO4 - 1.33g, NaCl - .5g and pH was modulated at 7. The basal medium was also enriched with 1mL of a trace element solution comprising 10 mg - MnCl2, 11mg - CoCl2, 15.7mg - CuSO4, 11.8 mg - NiCl2, 0.78g - CaCl2, 6.3 mg - CrCl2 and 0.97 g - FeCl3 per litre 13,12. The isolates were incubated in synthetic medium for further selection of isolated polyethylene-degrading bacterial strain. The synthetic medium contained K2HPO4-1g; (NH4)2SO4-1g; NaCl- 1g; MgSO4.7H2O- 0.5g; KH2PO4- 0.2g; FeSO4.7H2O- 0.01g; MnSO4.H2O- 0.001g; CuSO4.5H20- 0.001g; ZnSO4.7H2O- 0.001g; CaCl2.2H2O- 0.002g; polyethylene powder-332119 (Sigma-Aldrich) per 1000ml, pH adjusted to 7.2 14, 15, 16.
Identification of Polyethylene Degrading Strain
Automated Microbial Identification System
BIOMERIEUX VITEK 2 compact system version 07.01 was used for bacterial identification by biochemical interpretation using colorimetric reagent cards. VITEK 2 compact system had four colorimetric reagent cards i.e. Yeasts-like organisms, Gram negative fermenting bacilli & non-fermenting bacilli, Gram positive spore-forming bacilli and non-spore forming bacilli, Gram positive cocci & yeasts. Test results of an unknown organism were compared to the database of colorimetric reagent cards and to deduce a quantitative value for adjacency to every database taxa.
16S rRNA Sequence Analysis
Genomic DNA of degrading strain was extracted for 16SrRNA gene sequence examination. Polymerase chain reaction (PCR) was executed to amplify 16SrRNA region using the universal forward primer 5'-AGAGTTTGATCMTGGCTCAG-3' & the reverse primer 5' TACGGCTACCTTGTTACGACTT-3'. The amplification reaction was setup in 25µl PCR reaction including 5µl of isolated DNA, 1.5µl - forward primer & reverse primer each, 5µl - deionized water along with 12µl - Taq master mix (G-Biosciences). The amplification stage was performed for 35 cycles using a thermal cycler (BioRad, UK) as demonstrated: an initial denaturation on 95 °C up to 5 minute subsequently denaturation on 94 °C up to 30 seconds, annealing on 53 °C up to 1 minute, and 72 °C up to 2 minute with a last extension upto 10 minute at 72 °C. Amplified PCR products were checked by electrophoresis using 1.5% agarose gel and purified using Montage PCR clean up kit (Millipore). 16SrRNA sequencing was performed using ABI PRISM (Applied Biosystems). BLAST was performed to search any similarity between the extracted sequences and the NCBI database.
Degradation Analysis
Test Samples
The test samples were obtained through a random sampling process from the local market in Jaipur, India. Three polymeric film samples were collected and named as PE-S3, PE-S2 & PE-S1. These films were identified using an ATR-FTIR spectrophotometer spectrum (PerkinElmer Spectrum Version 10.4.00) and float/sink test in ethanol solution17. Samples were marked as PE-S1 (thickness- 19 microns) identified as high density polyethylene, PE-S2 (thickness- 10 microns) identified as low-density polyethylene and PE-S3 (thickness- 38 microns) identified as high density polyethylene.
Biodegradation Experiments
In-vitro biodegradation tests were performed on three different polyethylene film samples (one LDPE and two HDPE) in synthetic medium. Synthetic media (100 ml) were added to Erlenmeyer flasks. Polyethylene samples were dried at 60°C overnight and sterilized in 70% ethanol for 30 minutes. Each flask was inoculated with 10% of the selected degrading strain at log phase culture. Then inoculated flasks medium were incubated on a rotary shaker at an optimum temperature (35°C) for 30 days. After every 3 days, the growth of the degrading strain was checked using spectrophotometric analysis. The tests were conducted in triplicates.
Analytical Characterization for Monitoring Degradation
Spectroscopic Analysis
The experiments of biodegradation were setup with the inoculation of the degrading strain and control (without bacterial inoculation). Initial readings were taken for both control and bacterial inoculated medium, and it was counted as the day zero reading. Growth of the strain was assessed by determining the absorbance of the medium at 600 nm employing UV-Visible Spectrophotometer (Systronics 118) at 0, 3, 7, 10, 14, 17, 21, 24, 28 and 30 days.
Determination of Weight Loss
The film samples were recovered after incubation and washed with 2% Sodium Dodecyl Sulphate solution for 4 hours followed by distilled water to examine biodegradation of polyethylene film samples by isolated strain. The weight loss of each sample was calculated by comparing weight before & after treatment. The measure of polyethylene degradation by the bacteria was obtained with the weight difference of polyethylene samples between before and after the treatment. The average percentage weight loss of sample films was calculated as follows:
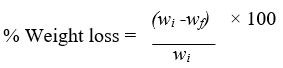
Here, wi - primary weight of film & wf - final weight of film.
FTIR (Fourier Transform Infrared Spectroscopy) Analysis
Nature of the functional group of the polyethylene changed due to bacterial exposure. These changes were analysed using FTIR spectroscopy (Shimadzu IR Affinity-1). Analysis was carried out in the range of 400-4000 cm-1 at ambient temperature over 32 scans per spectrum. The changes in Keto Carbonyl Index i.e. I1715/I1465 and Ester Carbonyl Index i.e. I1740/I1465 18 were measured and calculated in triplicate before and after bacterial incubation. These parameters were considered to assess the degradation of polyethylene.
Statistical Analysis
Statistical analysis was performed using R language version 3.4.319 for change in weight loss, Keto and Ester Carbonyl Index. Mixed Model ANOVA (analysis of variance) was performed to test significant interactions between different polyethylene film materials and there were either significant difference between the values before and after the degradation process. All treatments were done in triplicate.
Results and Discussion
Enrichment, Selection, and Identification of Polyethylene-Degrading Bacteria
A total of eight single colonies were isolated and labelled each consecutively (e.g. TN1, TN2, etc.). For further selection, all eight isolates were grown in synthetic medium with polyethylene powder as a carbon source. The isolate that showed maximum growth (maximum O.D value) were finally selected. Among these isolates, TN2 showed efficient potential growth (0.55 O.D.) on synthetic medium with polyethylene as a carbon source and was then selected for the biodegradation analysis. This mesophilic bacterium was Gram positive and rod shape. Its colonies were opaque, smooth and moist. The size of colonies on basal medium was 0.3-0.5mm. The optimum temperature and pH were 35°C and 7, respectively. This strain was thus further evaluated for biodegradation on both LDPE and HDPE thin films.
The VITEK 2 compact system was used for biochemical identification of the bacterial strain (TN2). The biochemical characterization is shown in Table 1.
Table 1: Biochemical characterization of the isolated Gram positive bacteria TN2 by VITEK2 compact system.
|
Well id |
Biochemical tests |
Test results |
|
12 |
Alanine ARYLAMIDASE (AlaA) |
+ |
|
15 |
Ala-Phe-Pro ARYLAMIDASE (APPA) |
(-) |
|
11 |
ALPHA-GALACTOSIDASE (AGAL) |
+ |
|
46 |
ALPHA-GLUCOSIDASE (AGLU) |
+ |
|
27 |
ALPHA-MANNOSIDASE (AMAN) |
- |
|
9 |
BETA-GALACTOSIDASE (BGAL) |
+ |
|
41 |
BETA-GLUCOSIDASE (BGLU) |
+ |
|
43 |
BETA-MANNOSIDASE (BMAN) |
- |
|
14 |
BETA-N-ACETYL-GLUCOSAMINIDASE (BNAG) |
(+) |
|
1 |
BETA-XYLOSIDASE (BXYL) |
+ |
|
18 |
CYCLODEXTRIN (CDEX) |
- |
|
19 |
D-GALACTOSE (dGAL) |
+ |
|
53 |
D-GLUCOSE (dGLU) |
- |
|
31 |
D-MANNITOL (d-MAN) |
+ |
|
32 |
D-MANNOSE (dMEN) |
+ |
|
34 |
D-MELEZITOSE (dMLZ) |
+ |
|
54 |
D- RIBOSE (dRIB) |
- |
|
47 |
D-TAGATOSE (dTAG) |
- |
|
48 |
D- TREHALOSE (dTRE) |
- |
|
25 |
ELLMAN (ELLM) |
- |
|
61 |
ESCULIN hydrolysis (ESC) |
+ |
|
30 |
Glycine ARYLAMIDASE (GlyA) |
- |
|
21 |
GLYCOGEN (GLYG) |
- |
|
58 |
GROWTH IN 6.5% NaCl |
- |
|
50 |
INULIN (INU) |
+ |
|
59 |
KANAMYCIN RESISTANCE (KAN) |
- |
|
4 |
L-Aspartate-ARYLAMIDASE (AspA) |
- |
|
5 |
Leucine- ARYLAMIDASE (LeuA) |
+ |
|
3 |
L-Lysine-ARYLAMIDASE (LysA) |
- |
|
8 |
L-Proline ARYLAMIDASE (ProA) |
- |
|
10 |
L-Pyrrolydonyl- ARYLAMIDASE (PyrA) |
+ |
|
39 |
L-RHAMNOSE (IRHA) |
- |
|
29 |
MALTOTRIOSE (MTE) |
+ |
|
24 |
METHYL-A-D-GLUCOPYRANOSIDE acidification |
- |
|
26 |
METHYL-D-XYLOSIDE (MdX) |
- |
|
22 |
myo-INOSITOL (INO) |
(+) |
|
36 |
N-ACETYL-D-GLUCOSAMINE (NAG) |
- |
|
60 |
OLEANDOMYCIN RESISTANCE (OLD) |
(+) |
|
37 |
PALATINOSE (PLE) |
+ |
|
7 |
Phenylalanine ARYLAMIDASE (PheA) |
+ |
|
44 |
PHOSHORYL CHOLINE (PHC) |
- |
|
63 |
POLYMIXIN_B RESISTANCE |
+ |
|
56 |
PUTRESCINE assimilation (PSCNa) |
- |
|
45 |
PYURVATE (PVATE) |
+ |
|
62 |
TETRAZOLIUM RED (TTZ) |
+ |
|
13 |
Tyrosine ARYLAMIDASE (TyrA) |
+ |
Here, + = Positive Result
- = Negative Result
(+) = Towards Positive
(-) = Towards Negative
For strain molecular identification, 16SrRNA gene sequencing was used. The NCBI BLAST database analysis showed that TN2 strain was closely related to Microbacterium genus as it was showing 99% similarity to Microbacterium barkeri. Consequently, the phenotypic and phylogenetic characteristics of the isolate TN2 made it a novel strain of M. barkeri (M. barkeri SH20) as polyethylene-utilizing bacteria which is not screen as far for biodegradation, and evaluation of its degrading capacity on polyethylene. Therefore, the nucleotide sequence was added in GeneBank NCBI as Microbacterium barkeri SH20 (Accession No. KY887791.1). To determine the taxonomic position of this strain, neighbour-joining phylogenetic tree (Figure 1) based on 16SrRNA sequencing was constructed (Mega7) showing nearest and related neighbours.
 |
Figure 1: Neighbour-joining phylogenetic tree showing strain TN2 derived from |
Growth Rate Monitoring of Bacterial Strain
The degradation ability of M.barkeri SH20 (TN2) bacterial strain was determined through treatment on one LDPE and two different HDPE films collected from the local market of Jaipur. The spectral growth was monitored for growth curve analysis of the bacterial isolate (Figure 2), and the degradation studies were conducted for 30 days. There was an increase of absorbance in the strain-inoculated treatment at 600nm. The absorbance of the PE-S3 HDPE films increased from 0.217 (± 0.015) to 0.420 (± 0.071), for PE-S2 LDPE from 0.215 (± 0.018) to 0.415 (± 0.022), and for PE-S1 HDPE films from 0.219 (± 0.02) to 0.392 (± 0.104) in comparison to control (without bacterial inoculation) which decreased from 0.092 (± 0.004) to 0.033 (± 0.002). In a report turbidity increased in mineral salt medium due to bacterial action on polyethylene20. In the present study, M.barkeri SH20 was able to grow in a medium containing only LDPE and HDPE films strips as carbon source. Thus, this strain had the potential to degrade polyethylene.
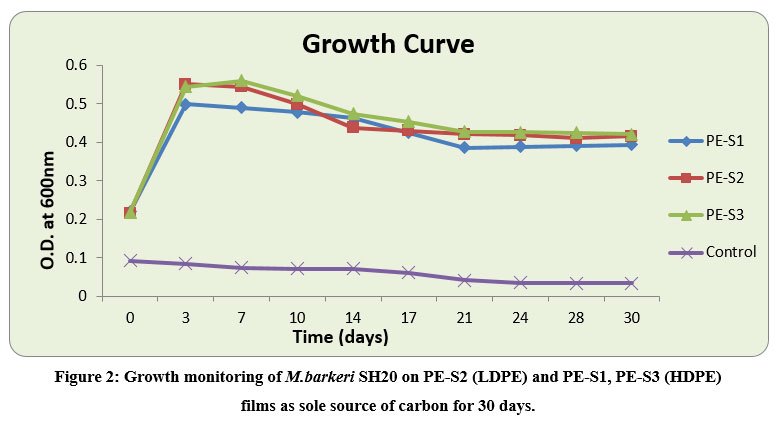 |
Figure 2: Growth monitoring of M.barkeri SH20 on PE-S2 (LDPE) and PE-S1, PE-S3 (HDPE) films as sole source of carbon for 30 days. |
Evaluation of Weight loss
The LDPE and HDPE film-treatments with M.barkeri SH20 showed weight losses after 30 days. The highest % weight loss of HDPE PE-S1 films were .985±0.23%, followed by 0.846±0.12% and 0.459±0.10% of PE-S2 and PE-S3, respectively (Figure 3). Results indicated that M.barkeri SH20 strain was effective in degrading both high density and low density polyethylene films. The ANOVA Mixed Model showed that there were significant differences in the weight loss values of both LDPE and HDPE samples between before and after bacterial treatment (p < 0.05).
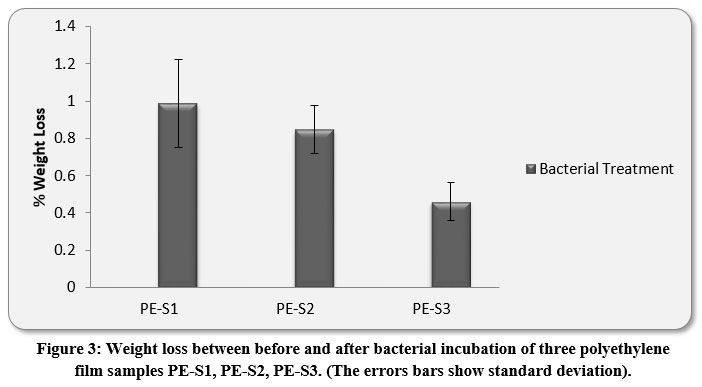 |
Figure 3: Weight loss between before and after bacterial incubation of three polyethylene film samples PE-S1, PE-S2, PE-S3. (The errors bars show standard deviation). |
FTIR studies
The Keto and Ester Carbonyl Index (KCBI and ECBI respectively) of 3 film samples was precisely analysed including control films to monitor the degradation (Figure 7). PE-S1 (HDPE) film sample showed increase in intensities of IR absorption in peaks (Figure 4). The HDPE film showed changes in the peak intensities and increases in IR peaks of C=O regions between 1870 to 1540 cm-1 which included carbonyls, esters, carboxylic acid, aldehydes etc. as compared to the control. For instance, a strong peak of carbon dioxide (2349cm-1) was detected during bacterial strain incubation. PE-S2 (LDPE) film samples treated with M.barkeri SH20 bacterial strain showed a sharp increase in IR absorption as compared to control samples (Figure 5). Additionally, the C=O stretching of both weak and strong peaks were observed in the range of 1870-1540 cm-1 which included carbonyl, esters, carboxylic acid, aldehydes functional groups etc. The SH20-treated PE-S3 (HDPE) films showed increase in IR absorptions as compared to the control samples (Figure 6). Also, changes in the C=O stretching peaks were observed in the1870 - 1500 cm-1 range and in absorption of peaks were observed between 2400-2000 cm-1 which included O=C=O stretching and C≡C stretching.
The amount of KCBI and ECBI of PE-S1 films were decreased from 19.05±0.31 to 12.50±1.2 and from 19.10±0.3 to 12.54±1.1 respectively. Furthermore, the rate of KCBI and ECBI of PE-S2 LDPE films were also decreased from 2.33±0.04 to 1.84±0.07 and from 2.28±0.1 to 1.92±0.03 respectively. For PE-S3, the amount of KCBI and ECBI was also decreased from 6.02±0.3 to 5.91±0.2 and from 6.14±0.3 to 5.66±0.3 after bacterial incubation for 30 days, respectively. Both Keto and Ester Carbonyl Indexes were highly significant (p-value < 0.01), which implied that bacterial treatment had significant interactions between different polyethylene film materials and there was significant differences between the values before and after the degradation process.
Figure 4, 5 and 6 showed new absorption bands in the range of 1700-1600 cm-1 apparently due to the generation of C=O groups generate at the bacterial attachment event on HDPE and LDPE films that initiated the biodegradation process21. Differences in Keto and Ester Carbonyl Index of before and after treatment could be due to changes in the C=O stretch and formation of aldehyde or ketone groups. Additionally, there were peaks at 1000-1760cm-1, possibly due to oxidation of –OH groups. Control samples for both LDPE and HDPE films did not show the presence of the previously mentioned functional groups.
Previous studies reported the decrease in carbonyl index after biotic exposure22, 23. Incubation with bacterial strain for 30 days showed depletion of amount of carbonyl residues and carbonyl index24. However, in a report25 an increase in Keto and Ester Carbonyl Index by 0.17±0.009 and 0.183±0.008 in 30 days during a degradation study of HDPE film treated with a fungal isolate, A.terreus MF12. Thus, in comparison to previous reports, M.barkeri SH20 had more potential to degrade significant amounts of LDPE and HDPE film.
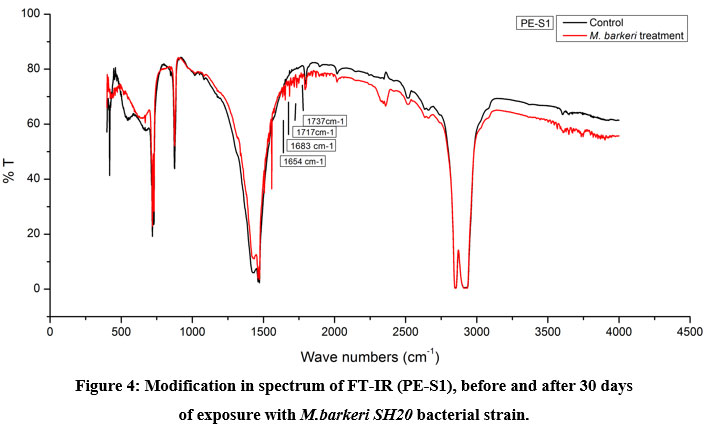 |
Figure 4: Modification in spectrum of FT-IR (PE-S1), before and after 30 days |
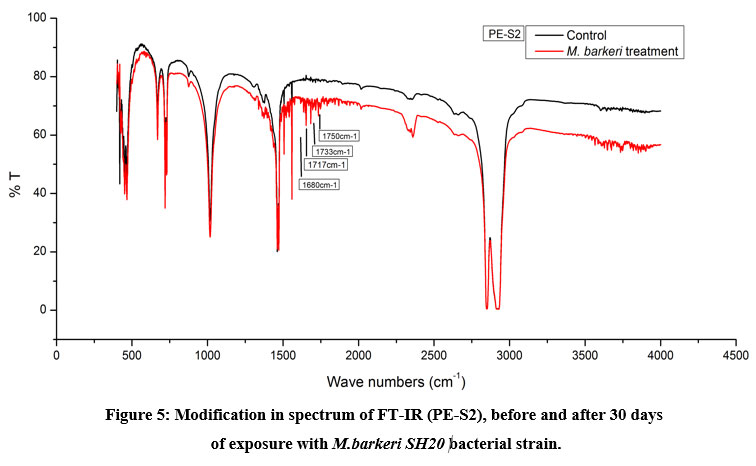 |
Figure 5: Modification in spectrum of FT-IR (PE-S2), before and after 30 days of exposure with M.barkeri SH20 bacterial strain. Click here to view Figure |
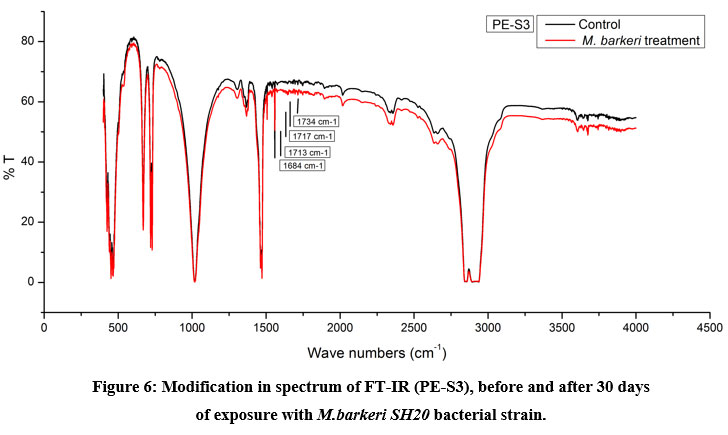 |
Figure 6: Modification in spectrum of FT-IR (PE-S3), before and after 30 days |
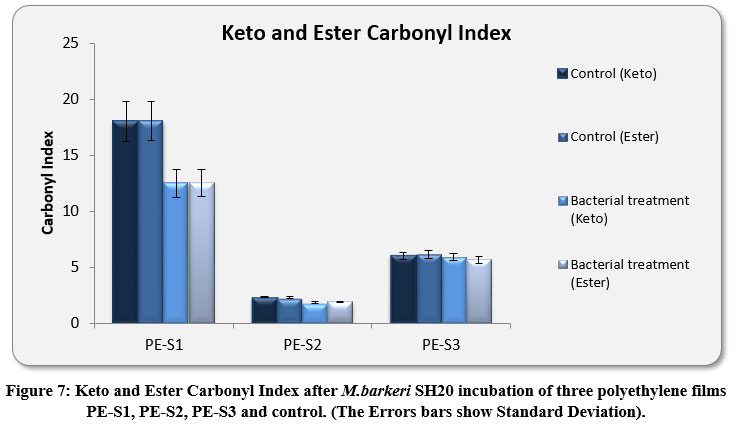 |
Figure 7: Keto and Ester Carbonyl Index after M.barkeri SH20 incubation of three polyethylene films PE-S1, PE-S2, PE-S3 and control. (The Errors bars show Standard Deviation). |
Conclusion
In the present study, bacterial isolate Microbacterium barkeri SH20, isolated from motor-oil contaminated soil (TN2), is capable of adhering on surface and utilize both low and high density polyethylene films as carbon source efficiently. The biochemical characterization of isolated strain was based on automated microbial identification system. TN2 was identified through 16SrRNA gene sequencing and strain was closely related to Microbacterium genus. It was identified as Microbacterium barkeri species and the nucleotide sequence has been deposited in gene bank as Microbacterium barkeri SH20 (Accession No. KY887791.1).
The degradation evidences of bacterial isolate has been validated by gravimetric weight loss, modification carbonyl-index through Fourier Transform Infrared Spectroscopy analysis and evolution of different surface chemical moieties by microbial colonization. Significant growth rate was observed with M. barkeri SH20 on all three different polyethylene film samples as compare with corresponding control. Degradation of three samples (PE-S1, PE-S2 and PE-S3) demonstrates that, PE-S1 HDPE films can degrade at highest rate followed by PE-S2 LDPE films with M. barkeri SH20. Thus, M. barkeri SH20 showed capacity of degradation of both LDPE and HDPE films.
Some studies reported the use of Microbacterium in consortium with other microorganisms associated with the LDPE degradation 26, 27. However, to our knowledge, this is the first time that Microbacterium barkeri SH20 is reported as a novel strain isolated from motor-oil contaminated soil, showing significant growth and biodegradation on polyethylene matrices. Thus, this microorganism can be useful in landfills containing solid plastic waste.
Acknowledgment
The authors are thankful to CAS, Department of Zoology, University of Rajasthan (UOR); USIC (University Science Instrumentation Centre), UOR; MNIT (Malviya National Institute of Technology), Jaipur for giving technical support.
Conflict of Interest
The authors proclaim that there is no conflict of interest with respect to publication of this paper.
Funding Source
The authors are thankful for financial support from CAS (Centre for Advance Studies), Department of Zoology, University of Rajasthan and UGC-BSR fellowship.
References
- Nowak B, Pajak J, Drozd-Bratkowicz M, Rymarz G. Microorganisms participating in the biodegradation of modified polyethylene films in different soils under laboratory conditions. Int Biodeterior Biodegrad. 2011; 65:757-767.
CrossRef - Sheik S, Chandrashekar K.R, Swaroop K., Somashekarappa H M. Biodegradation of gamma irradiated low density polyethylene and poly propylene by endophytic fungi. Int. Biodeterior. Biodegr. 2015; 105, 21-29.
CrossRef - Geyer R, Jambeck J R, Law K R. Production, use, and fate of all plastics ever made. Sci Adv. 2017; 3(7): e1700782.
CrossRef - Central Pollution Control Board Annual Report 2016-17. PR Division, Ministry of Environment & Forests and Climate Change, Govt. of India. 2017. (http://cpcb.nic.in/uploads/plasticwaste/Annual_Report_2016-17_PWM.pdf).
- Shrivastava RS. Our environment: challenges and solutions. Diamond Pocket Books Pvt Ltd. 2016.
- Shah A.A., Hasan F., Hameed A., Ahmed S. Biological degradation of plastics: a comprehensive review. Biotechnology Advances. 2008; 26: 246-265.
CrossRef - Kowalczyk, A., Chyc, M., Ryszka, P., & Latowski, D. Achromobacter xylosoxidans as a new microorganism strain colonizing high-density polyethylene as a key step to its biodegradation. Environmental Science and Pollution Research. 2016; 23(11): 11349-11356.
CrossRef - Lu, J. C., Li. Z. T., Hussain, K. and Yang, G.K. Middle East J Sci Res. 2011; 7: 738-740.
- Restrepo-Florez JM, Bassi A, Thompson MR. Microbial degradation and deterioration of polyethylene – A review. Int Biodeterior Biodegrad. 2014; 88:83-90.
CrossRef - Liu H, Yao J, Yuan Z, Shang Y, Huilun C, Wang F, Masakorala K, Yu C, Cai M, Blake RE, Choi MMF. Isolation and characterization of crude-oil-degrading bacteria from oil-water mixture in Dagang oilfield, China. Int Biodeterior Biodegrad. 2014; 87:52-59.
CrossRef - Chebbi A, Hentati d, Zaghden H, Baccar N, Rezgui F, Chalbi M, Sayadi S, Chamkham. Polycyclic aromatic hydrocarbon by a newly isolated Pseudomona so. Strain from used motor oil-contaminated soil. Int Biodeterior Biodegrad. 2017; 122:128-140.
CrossRef - Yoon MG, Jeon HJ, Kim MN. Biodegradation of polyethylene by a Soil bacterium and AlkB cloned Recombinant Cell. J Bioremed Biodegrad. 2012; 3:1000145.
- Kim MN, Park T. Degradation of Poly (L-lactide) by a Mesophilic Bacterium. J Appl Polym Sci. 2010; 117:67-74.
CrossRef - Kavitha, R., Mohanan A. K., and Bhuvaneswari V. Biodegradation of low density polyethylene by bacteria isolated from oil contaminated soil. International Journal of Plant, Animal and Environmental sciences. 2014; 4(3): 601-610.
- Abraham J., Ghosh E., Mukherjee P., and Gajendiran A. Microbial degradation of low density polyethylene. Environmental Progress & Sustainable Energy. 2017; 36(1): 147-154.
CrossRef - Skariyachan S., Setlur A. S., Naik S. Y., Naik A. A., Usharani M., and Vasist, K. S. Enhanced biodegradation of low and high-density polyethylene by novel bacterial consortia formulated from plastic-contaminated cow dung under thermophilic conditions. Environmental Science and Pollution Research. 2017; 24(9), 8443-8457.
CrossRef - Jung, M. R., Horgen, F. D., Orski, S. V., Rodriguez, V., Beers, K. L., Balazs, G. H., & Hyrenbach, K. D. Validation of ATR FT-IR to identify polymers of plastic marine debris, including those ingested by marine organisms. Marine pollution bulletin. 2018; 127: 704-716.
CrossRef - Artham T., Sudhakar M., Venkatesan R., Madhavan N. C., Murty K., and Doble M. Int. Biodeterior. Biodegrad. 2009; 63: 884-890.
CrossRef - R Core Team. R. A language and environment for statistical computing. R Foundation for Statistical Computing,Vienna, Austria. 2017. ( https://www.R-project.org/.).
- Chatterjee S., Roy B., Roy D. and Banerjee R. Enzyme-mediated biodegradation of heat treated commercial polyethylene by Staphylococcal species, Polym. Degrad. Stabil. 2010; 95: 195-200.
CrossRef - Corti A, Muniyasamy S, Vitali M, Imama SH, Chiellini E. Oxidation and biodegradation of polyethylene films containing pro-oxidant additives: synergistic effects on sunlight exposure, thermal aging and fungal biodegradation. Polym Degrad Stabil. 2010; 5:1106-1114.
CrossRef - Roy PK, Titus S, Surekha P, Tulsi E, Deshmukh C, Rajagopal C. Degradation of abiotically aged LDPE films containing pro-oxidant by bacterial consortium. Polym Degrad Stab. 2008; 93:1917-1922.
CrossRef - Gilan I, Hadar Y, Sivan A. Colonization, Biofilm formation and biodegradation of polyethylene by a strain of Rhododcoccus ruber. Appl Microbiol Biotechnol. 2004; 65:97-104.
CrossRef - Hadad D, Geresh S, Sivan A. Biodegradation of polyethylene by thermophilic bacterium Brevibacillus borstelensis. J Appl Microbiol. 2005; 98:1093-1100.
CrossRef - Balasubramanian V., Natarajan K., Rajeshkannan V., & Perumal P. Enhancement of in vitro high-density polyethylene (HDPE) degradation by physical, chemical, and biological treatments. Environmental Science and Pollution Research. 2014; 21(21): 12549-12562.
CrossRef - Negi H, Gupta S, Zaidi MGH, Goel R Studies on biodegradation of LDPE film in the presence of potential bacterial consortia enriched soil. Biologija. 2011; 57(4):141–147.
CrossRef - Rajandas H, Parimannan S, Sathasivam K, Ravichandran M, Su Yin L. A novel FTIR- ATR spectroscopy based technique for estimation of low density polyethylene biodegradation. Polym Test. 2012; 31:1094-1099.
CrossRef






