Rhizobium Leguminosarum: A Model Arsenic Resistant, Arsenite Oxidizing Bacterium Possessing Plant Growth Promoting Attributes
Aritri Laha1,2
*
 , Somnath Bhattacharyya2
, Somnath Bhattacharyya2
 , Sudip Sengupta3
, Sudip Sengupta3
 , Kallol Bhattacharyya3
, Kallol Bhattacharyya3
 and Sanjoy GuhaRoy1
and Sanjoy GuhaRoy1
Corresponding author Email: lahaaritri@gmail.com
DOI: http://dx.doi.org/10.12944/CWE.16.1.09
Copy the following to cite this article:
Laha A, Bhattacharyya S, Sengupta S, Bhattacharyya K, GuhaRoy S. Rhizobium Leguminosarum: A Model Arsenic Resistant, Arsenite Oxidizing Bacterium Possessing Plant Growth Promoting Attributes. Curr World Environ 2021;16(1). DOI:http://dx.doi.org/10.12944/CWE.16.1.09
Copy the following to cite this URL:
Laha A, Bhattacharyya S, Sengupta S, Bhattacharyya K, GuhaRoy S. Rhizobium Leguminosarum: A Model Arsenic Resistant, Arsenite Oxidizing Bacterium Possessing Plant Growth Promoting Attributes. Curr World Environ 2021;16(1). Available From : https://bit.ly/3cV9B8s
Download article (pdf)
Citation Manager
Publish History
Select type of program for download
| Endnote EndNote format (Mac & Win) | |
| Reference Manager Ris format (Win only) | |
| Procite Ris format (Win only) | |
| Medlars Format | |
| RefWorks Format RefWorks format (Mac & Win) | |
| BibTex Format BibTex format (Mac & Win) |
Article Publishing History
| Received: | 14-10-2020 |
|---|---|
| Accepted: | 10-02-2021 |
| Reviewed by: | 
 Aparna B. Gunjal
Aparna B. Gunjal
|
| Second Review by: |

 Ladan Bayat
Ladan Bayat
|
| Final Approval by: | Dr. Mohammad Oves |
Introduction
The human populations of eastern India (West Bengal mainly) and Bangladesh are severely suffered by arsenic (As) contamination of water and food1, 2. The concurrent application of As enriched irrigation seems to be the major reason for soil build-up3, 4and subsequent accumulation in standing crops5. Generally, As residues are found in the top layer of soil or surface soil and enter the plants easily because of their low volatility and low solubility6. The As has both organic and inorganic forms7that found as an oxyanion in the environment 8. The high-risk involvement and high cost of remediation9emphasize the scope of bioremediation through novel As-tolerant microbes10.
In As polluted area, there are several soil-based microbes11with adaptive strategies for surviving in a contaminated atmosphere; mostly byutilizing the toxic arsenic as a nutrient for their metabolisms and proliferation6.Some bacteria such as Burkholderia12, Agrobacterium, Bacillus, Rhizobium are best suitable candidate for arsenic bioremediation and plant growth promotion (PGP)13.These microorganisms can have a dual purpose environmental beneficial impacts by metal decontamination and plant growth promoters14, 15. These PGP bacteria are able to enrich soil nutrient and able to mitigate soil arsenic also13.The physical and chemical process for arsenic mitigation are very costly so these rhizobium-legume symbiosis may a alternative new tool for As mitigation16.
The unique attributes of As resistance and plant growth promotion (PGP) in rhizospheric microbes is quite noteworthy. These adopted indigenous soil microbes tend to manifest several PGP traits through secretion of 1-aminocyclopropane-1-carboxylate (ACC), deaminase, indole-3-acetic acid (IAA), phosphate solubilizers, producing siderophores that reduce metal toxicity and encourage plant-assisted bioremediation17, 18, 19.Most remarkably, such PGP characters remain active even under intense arsenic stress conditions20, 21.
In this backdrop, we have categorized the study and identified efficient As-resistant bacterium from contaminated rhizosphericsoil, and investigated the PGP attributes of identified bacteria with a view to harvest their capacity to develop plant’s resistance to stress conditions, encourage plant growth, and give a path to contribute accelerated remediation of arsenic polluted soils.
Materials and Methods
Soil Sample Analysis
Soil samples (2 cm diameter, 10 cm depth) were collected in sterilized zipper bags to maintain an aseptic condition from arsenic (As) contaminated rhizospheric zone of lentil in Chakdaha, West Bengal (23º05' N latitude and 88º54' E longitude), India, that noted for As concentrations in the groundwater above World Health Organization (WHO)-defined safe limit22. Atomic AbsorptionaSpectrophotometera (AAS, Model-Perkin Elmer A Analyst 200) was used to measure total23 and available As24 of the soil samples.
Isolation;of;As-Tolerant;Bacteria
2 g of soil;(taken from lentil rhizosphere separately) was; suspended in 2 mL;sterile;distilled water, 1mL of each was re-suspended in Yeast Extract Mannitol (YEM) liquid medium separately in two conical flasks, spiked with 1 mM arsenate and 1 mM arsenate.The total experimental set up was incubated for 48 hours at 30°C25.After incubation, 2 ml bacterial culture was further inoculated in a YEM liquid medium.The procedure was repeated twice. Around 0.1;mL;of As spiked culture was spread on a YEMA (Yeast;Extract;Mannitol Agar) solid medium plate to isolate the As tolerant bacteria. To test the Rhizobium strain, the congo red YEMA test was categorized26.
MIC (Minimum Inhibitory Concentration) Value of the Bacteria
The MIC is the lowest concentration of arsenate or arsenite which entirely inhibits microbial growth and activity27, 28. 1.0 mL of overnight bacterial culture was inoculated in two conical flasks containing 99.0 mL of YEM liquid medium, containing either arsenite (NaAsO2; 1-50 mM) or arsenate (Na2HAsO4·7H2O; 1-500 mM) separately and;incubated;at 30ºC with 48 hrs of shaking. The OD (optical density, measurement of microbial growth) of the bacterial cultures was detected using a UV-Vis spectrophotometer (Model: Varian CARY-50) at 600 nm wavelength.
Identification of the As;Tolerant;Bacteria
The As;tolerant;bacterial;genomic DNA was;isolated and PCR amplification for molecular identification by 16S rRNA sequence analysis27.The bacterial isolate was studied for Gram reaction, colony morphology and characterized for catalase, urease, and oxidase activities by standard protocols29.
Scanning Electron Microscopic (SEM) Study
For the SEM study,First the bacterial cells were collected andallowed to wash properly with sodium;phosphate;buffer;(pH 7.4).Then a thin smear of bacterial cells were kept on a glass cover slip and prepared a smear, after that it was allowed to heat-fixover a flame;for;1–2 sec followed;by;fixation;with;2.5%;glutaraldehyde;for;45 min30. The slides;were;then;dehydrated with;50–90%;of;alcohol;solutions;and;finally;through;absolute;alcohol;for;5;min;each;and observed;under;a;15kV;scanning;electron;microscope;(HITACHI,;S-530,;SEM,;and;ELKO Engineering).
Arsenite Transformation; and Species Detection
The arenite; transformation; ability of the bacterial; isolate; was determined by modified laboratory-based standard protocol27. As concentrations were determined by using a spectrophotometric method31. The recoveries of the As species fromthe liquid culture medium by the efficient bacterial strains were determined by HPLC-ICP-MS32. All chemicals used for speciation study were reagent grade. All of the experimental solutions were prepared with Mili-Q (Millipore, Bedford, MA, USA) water. A Perkin Elmer Series 200 Micro Pump was used instead of the quaternary pump for the isocratic method. The isocratic mobile phase was 30 mM NH4H2PO4 at pH 5.6. The flow rate was 1.0 mL min-1 with 100 μL sample injections. The LC column effluent was directly attached to the nebulizer with PEEK tubing (1.59 mm OD) and a low dead volume PEEK connector (Part No.:WE024375).The isocratic mobile phase was 30 mM NH4H2PO4 at pH 5.6. The flow rate was 1.0 mLmin-1 with 100 μL sample injections. The LC column effluent was directly attached to the nebulizer with PEEK tubing (1.59 mm OD) and a low dead volume PEEK connector (Part No.: WE024375).
Plant Growth Promoting (PGP) Characteristics of the As Tolerant Bacterium
The isolated arsenic resistant bacterial PGP attributes (IAA-production,;ACC;Deaminase activity,;phosphate;solubilization,;nodulation,;and;siderophore;production) were determined in vitro in culture medium under arsenic stress conditions (spiking levels being 0 mg/L,;15 mg/L,;and 30 mg/L;As(III)/As(V)).
Ability to Solubilize Phosphate
This encompassesthe growth of the bacterial strain in Pikovskaya’s medium33(containing 0.5%;of;tricalcium;phosphate;(TCP);spiked with three levels of arsenate and arsenite 0, 15, 30 mg/kg) at 30°C for 5-6 days and 170 rev/min. Then the culture was allowed to centrifuge at 6500 times gravity (×g) and supernatant was collected. The solubilized phosphate in supernatant of culture medium was estimated34.
Screening of Indole Acetic Acid (IAA)
The As resistant isolates were determined by growing them in an L-tryptophan (0.5 mg/ml) supplemented minimal medium;in;presence;of;a different;concentration;of As (0, 15, 30 mg/L) and incubated for 5 days in the dark at 30 0C. 2 ml bacterial suspension was transferred in 100 µl 10 mM orthophosphoric acid and 4 ml Salkowski’s reagent (2% solution of 0.5 M FeCl3 in 35% perchloric acid) in a test tube. The entire mixture was shaken vigorouslybefore incubation for 45 min until a pink colour develops. Absorbance of the resultant solution was determined at 530 nm for obtaining the content of IAA-like molecules in liquid culture medium35.
ACC Deaminase Activity
The ACC Deaminase enzymeactivity is the quantity of α-ketobutyric acid production by the breakdown of ACC36 by the bacterial strain.To determine this, a minimal medium was prepared using 1-aminocyclopropane-1-carboxylic acid or ACC (3 gL−1) as a source of nitrogen, spiked with three different concentrations of As(V) and As(III) (0, 15, 30 mg/L) separately and the bacterial cells were grown. The quantity of ketobutyrate (KB) formed per mg of protein per hour is the total value of the specific enzyme activity.
Nodulation Efficiency
The nodulation efficiency of bacterial strain37 was assessed through a pot study. Soils were sterilized and the seeds of lentil were sown in the sterilized soil spiked with As (0, 15, 30 mg/kg) and the nodule counts were taken after 30 days.
Screening for Siderophore Production
The siderophores production was qualitatively assayed by using the Chrome Azural S method of Schwyn and Neilands35. The MM9 (Tris buffer, casamino acids (0.3%), L-glutamic acid (0.05%), (+)- biotin (0.5 ppm), and sucrose (0.2%)) liquid medium with the absence of Fe was used for bacterial growth. The bacterial culture medium was inoculated in this medium and allowed to incubate for 5 days at 30ºC temperature. For control 0.2 µM of iron (freshly prepared, filter-sterilized FeSO4.7H2O stock solution) was also inoculated. The stationary phase of bacterial culture wascollected and pelleted by centrifugation (6,500 x g for 15 minutes). In supernatant solution, the most important qualitative conformation of the presence of siderophore is simply the color change from blue to orange.
Statistical Analysis
Statistical computations were performed using Microsoft Excel 2016 and SPSS version 23.0.
Results
Characterization of the Experimental Site
The total As concentration and available As concentration of experimental rhizospheric soil of lentil were measured. Results revealed a considerable load of the total and available arsenic i.e. 17.2 ± 1.72 and 1.50 ± 0.27 mg kg-1 respectively (presented as the mean of three observations ± SD).
Isolation and Identification of the As Tolerant Bacteria
For isolation of Rhizobium, the YEMA medium with congo red was used.A few distinct colonies were found (Fig 1). From these,a distinct colony was picked and allowed to further study.
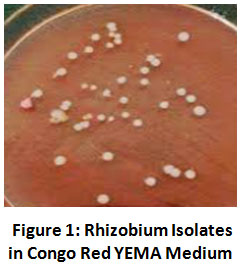 |
Figure 1: Rhizobium Isolates in Congo Red YEMA Medium. Click here to view Figure |
The isolated strains of rhizobacteria were found to be gram-negative and rod-shaped. Theseisolated strainswere studied for phenotypic and biochemical studieshave been represented in Table-1.The bacterial isolate waspositive for catalase, glucose, sucrose, sorbitol, lactose, mannitol, maltose, xylose, and fructose; while oxidase,indole, citrate, MR, VP, urease activity were found to be negative.
Table 1: Morphological and Biochemical Properties of the Bacteria.
|
Biochemical Tests |
Performance |
|
Catalase |
+ |
|
Oxidase |
- |
|
Urease |
- |
|
Indole |
- |
|
Citrate utilization |
- |
|
MR |
- |
|
VP |
- |
|
Fructose..++ |
++ |
|
Mannitol..++ |
++ |
|
Sorbitol..++ |
++ |
|
Lactose..++ |
++ |
|
Sucrose..++ |
++ |
|
Xylose..++ |
++ |
|
Maltose..++ |
++ |
|
Glucose..++ |
++ |
Further employing 16S rRNA gene sequencing a phylogenetic tree was prepared (Fig-2). These identified bacterial isolates are thus assumed to be Rhizobium leguminosarum (LAR-7, accession numberMK696942).
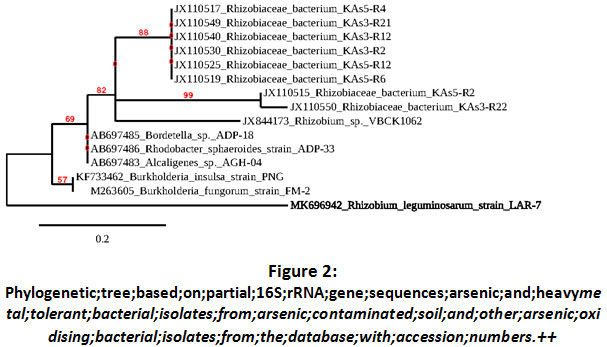 |
Figure 2: Click here to view Figure |
Further to confirm the results, the SEM study of the bacterial isolates (Fig- 3) was also categorized.
 |
Figure 3: SEM Image of the Bacterial Strain. Click here to view Figure |
MIC of the Arsenic Resistant Bacterial Isolates
The arsenic tolerant strains were tested for their minimum inhibitory concentration of As.The result showed that LAR-7(Rhizobium leguminosarum) hada high MIC value.Bacterial isolate had emerged with a high MIC towards arsenate (260 mM) and arsenite (27.5 mM).
Arsenite’transformation’and’species’detection
The As accumulation and volatilization potential of the isolate wasexperimented with through a quantitative estimation by incubating for 24 hr in a liquid culture medium spiked with 30mM As(III). The As content in liquid medium wasanalyzedto attain the quantity of Asdecontamination. The bacterial strain LAR-7 has shown a significantlevel of arsenic decontamination ability. 35% of total arsenic was decreased from the liquid culture medium containing 30 mMof arsenite.The bacterial transformation of arsenite to arsenate was 437.5μM h-1, which is considered 35%of the total arsenite of media (Table 2). LAR-7 could oxidize 35% of 30 mM arsenite within 24 hours.
Table 2: Arsenic Transformation and Species Detection of the;bacterial’isolates’under 10mg/L As(III);Application.
|
Isolate |
Total As |
Arsenite As(III)++ |
Arsenate As(V)++ |
% transformation As(III)+ to As(V)+ |
Species recovery |
|
LAR-7 |
9.32±2.21 |
5.69±1.96 |
3.63±1.63 |
39% |
61% |
|
Control |
9.37±2.38 |
9.30±2.64 |
0.07±0.02 |
0% |
0% |
Each value is a mean ofthree replicates. mean ± standard deviation.
In this study, about 61% of the total As was recovered in As species. The transformation of arsenite to arsenate in liquid culture broth varied from 0 (uninoculated control) to 39% (medium inoculated with LAR-7) by the selected arsenic resistant microbial strains.
Potential Plant; growth;promoting;properties;of;As tolerant bacteria
The plant;growth;promoting;traits;of;the;As;tolerant;isolate was categorized. The strain LAR-7 was able to solubilize phosphate, produce IAA and ACC Deaminase under As(V) and As (III) stressed conditions (Table-3).
Table 3: Plant Growth Promoting Properties of the Bacterium LAR-7.
|
Arsenic species |
As spike (mg/kg) |
Phosphate solubilization (µgL-1) |
IAA Production++ (μM IAA ml-1) |
Acc deaminase’activity (nM α-ketobutyrate mg protein-1h-1)== |
Number of nodule production |
Siderophore production |
|
As(III) |
0 |
440.8±53.01 |
16.4±8.62 |
1.99±0.56 |
112±22.3 |
+ |
|
15 |
140.0±26.35 |
10.0±5.32 |
1.09±0.62 |
90±21.3 |
+ |
|
|
30 |
137.2±23.69 |
10.0±5.26 |
1.00±0.29 |
90±13.5 |
+ |
|
|
As(V) |
0 |
440.8±51.06 |
16.4±7.69 |
1.99±0.45 |
112±22.9 |
+ |
|
15 |
440.5±22.35 |
12.8±6.23 |
1.99±0.68 |
110±12.6 |
+ |
|
|
30 |
337.5±21.05 |
12.8±6.12 |
1.87±0.64 |
100±11.6 |
+ |
Each;value;is’a’mean’of’three’replicates. mean ±’standard;deviation.
LAR-7 was observed to solubilize a good amount of phosphate (440.8, 440.5, 337.5μg/L when the medium was spiked with 0,15,30 mg/kg arsenate respectively).Similarly, the same results were also found under arsenite stress conditions. This bacterial isolate also solubilizesphosphate 440.8, 140.0, 137.2 μg/L when the medium was spiked with 0,15,30 mg/kg arsenite respectively. Under As stress condition, theamount of phosphate solubilization was decreased but still the bacteria also able to solubilize phosphate.
LAR-7 also was’able’to’produce;Indole;acetic;acid;(IAA) and had a good ACC Deaminase activity. The quantity of IAA and ACC Deaminase activity were 16.4,12.8,12.8 μM IAA ml-1(medium containing 0,15,30 mg/kg arsenate)and 1.99,1.99,1.87 nM α-ketobutyrate mg protein-1h-1 (medium containing 0,15,30 mg/kg arsenite) respectively. Under arsenite (medium containing 0,15,30 mg/kg arsenite) stress conditionthe quantity of IAA and ACC Deaminase activity were 16.4,10.0,10.0 μM IAA ml-1 and 1.99,1.09,1.00 nM α-ketobutyrate mg protein-1h-1 respectively. Under arsenite and arsenate stress condition arsenic resistant bacterial isolate LAR-7 was also able to produce the root nodule and also the ability to produce siderophore.The siderophore production was qualitatively tested. So LAR-7 (Rhizobium leguminosarum) is an arsenic resistant bacterium also able to carry a bunch of plant growth promoting properties (Fig- 4).
 |
Figure 4: PGP Attributes of the Bacterial Strain LAR-7. Click here to view Figure |
Discussion
Identification and isolation;of;As-resistant;bacterial;strains;from;contaminated;soils
In the course of identification and;characterization;of;arsenic;resistant;PGP;bacteria;from the As polluted area in the present investigation LAR-7 (Rhizobium leguminosarum ) bacterial isolate had emerged with a high MIC towards arsenate (260 mM) and arsenite(27.5) mM which is higher than the previously reported MIC for arsenate (260 mM) and arsenite (27.5mM) for Rhizobium radiobacterof 10 mM of As(V) in agricultural soils38 and also greater than other As-oxidizing, As-resistant bacteria in soil;(183 mM arsenate and 6 mM of arsenite)15, in ground water (200 mM;arsenate and 5 mM;arsenite)39, in mines (10 mM As(V))40. LAR-7 also showed a greater MIC value than Rhizobium radiobacter which showed the highest;MIC;of >1,500 mg/L;of arsenic41.The isolate LAR-7 in our present;investigation;was;identified;as Rhizobium leguminosarum (based;on;16S;rRNA;gene;sequence;homology) which can oxidize at the rate of 437.5μM h−1 (35%) in presence of an arsenite concentration (30 mM) within 24 h under laboratory condition. The behaviour is somewhat similar to Alcaligenes sp. completely oxidizes arsenite within 40 h when exposed to relatively reduced arsenite concentrations (1 mM)42. The enhanced oxidation efficiencies of isolates in the present research have been quantitatively corroborated via speciation analysis. It was previously reported that some beta and gamma proteobacteria are also able to oxidise arsenic43.
Plant Growth Promoting Attributes in As Resistant, Arsenite Oxidizing Bacterial Strains
Recent studies have thrown some light on the PGP traits shown by the As oxidizing bacteria. The isolated bacterial strains of Acinetobacter sp., Klebsiella sp., Pseudomonas sp., Enterobacter sp. and Comamonas sp. from As- polluted tannery wastes under agricultural lands in Thailand35 possess both As tolerance and siderophore production capacity10. Arthrobacterglobiformis,:Bacillus:megaterium,:Bacillus:cereus,:Bacillus;pumilus,:Staphylococcus:lentus,Enterobacter;asburiae,;Sphingomonas;paucimobilis,:Pantoea:spp.,”Rhizobium:rhizogenes,and:Rhizobium:radiobacter:are:some:arsenic:resistant:bacteria.Rhizobium:rhizogenes:has:ahigh:MIC:Value13.Rhizobium;leguminosarum;are;also;capable;to;mitigate;As44.
Rhizobium community can survive the heavy metal pollutant soil and effective to enrich the soil;nutrient;and;save;the;enhance;the;growth;of;the;plant;in;contaminated;field16.Bradyrhizobium japonicum:has:potential:as:a:plant:growth-promoting:rhizobacterium also carries a bunch of PGPR attributes. Rhizobium melilotialso produce IAA-like molecules and able to solubilize a significant amount of phosphate45 under stress conditions.Rhizobium japonicum, Rhizobium spp, Rhizobium leguminosarum are the three plant growth promoting bacteria which are showed their PGPR activities under heavy metal stress conditions. The LAR-7 also could solubilize a significant amount of phosphate and produce IAA. Most bacteria under:Pseudomonas:sp.,:Acinetobacter sp.:and:Paenibacillus sp.;were reported to be potential plant growth promoters35 along withBacillus aryabhattaiwhich behaves similar to our studied strain forbeing an As resistant plant growth promoter20.
Similar observations were also obtained with As-resistant bacteria of Alpha proteobacteria, Beta proteobacteria, and Gamma proteobacteria manifesting potential PGP attributes 46, 10. Pseudomonas sp. has been reported to possess high As(III) oxidizing capacity while at the same time found to solubilize a significant amount of phosphate, indulge in siderophores,:IAA-like molecules,:and:ACC:Deaminase:production35.
The:present investigation has indisputably established the manifestation of:PGP:traits:ofAs:tolerant, As;oxidizing;bacterial;isolate;Rhizobium leguminosarum, insolubilizing phosphate;, producing siderophores, root nodule, IAA-like molecules, and ACC deaminase under As stress. Rhizobium leguminosarum (LAR-7) had emerged as best performing candidate isolates with regard to As resistance and PGP traits.
Conclusion
In pursuit of providing environmental safeguard to restore food safety and sustaining food security to the burgeoning population and combat abiotic pollution, a low-cost alternative to exorbitant pollution control strategies remained an absolute priority. The outcome of the present investigation revealed the candidate bacterial isolate Rhizobium leguminosarum as a potent, novel bacterial strain for As mitigation and may be useful, through fulfillment of mass production and field validation protocols, in As decontamination and plant growth promotion.
Acknowledgement
The authors are grateful to the ICAR-Niche area of Excellence-As Research Laboratory, Bidhan Chandra Krishi Viswavidyalaya (BCKV), Kalyani, Nadia as well as Department of Genetics & Plant Breeding and Department of Agronomy, Bidhan Chandra Krishi Viswavidyalaya (BCKV), Mohanpur, Nadia, West Bengal, India for providing technical and all other necessary assistance during the study.
Funding Source
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work.
References
- Chakraborti, D., Rahman, M.M., Mukherjee, A., Alauddin, M., Hassan, M., Dutta, R.N., Pati, S., Mukherjee, S.C., Roy, S., Quamruzzman, Q. and Rahman, M. Groundwater arsenic contamination in Bangladesh—21 Years of research. Journal of Trace Elements in Medicine and Biology, 2015: 31, 237-248.
CrossRef - Bhattacharyya, K. and Sengupta, S. Arsenic management options in soil-plant-food chain. In: Proceedings of the National Webinar On Arsenic Mitigation: A Nexus Approach; (Prasad Bishun D., Mandal Jajati., Kumar Sunil., Sohane R K, Eds.), 2020. pp. 17-23.
- Abedin, M. J., Cotter-Howells, J., and Meharg, A. A. Arsenic uptake and accumulation in rice (Oryza sativa L.) irrigated with contaminated water. Plant and soil, 2002: 240(2), 311-319.
CrossRef - Meharg, A. A., and Rahman, M. M. Arsenic contamination of Bangladesh paddy field soils: implications for rice contribution to arsenic consumption. Environmental science & technology, 2003:37(2), 229-234.
CrossRef - Bogdan, K. and Schenk, M.K. Evaluation of soil characteristics potentially affecting As concentration in paddy rice (Oryza sativa L.). Environmental Pollution, 2009: 157, 2617–2621.
CrossRef - Das, S., Jean, J. S., Kar, S., and Liu, C. C. Changes in bacterial community structure and abundance in agricultural soils under varying levels of As contamination. Geomicrobiology Journal, 2013. 30(7), 635-644.
CrossRef - Matschullat, J. As in the geosphere - a review. The Science of the Total Environment, 2000. 249, 297-312.
CrossRef - Frankenberger, W. T. Jr. and Arshad, M. Volatilization of As. In: Frankenberger, W.T. Jr. (Ed.). Environmental chemistry of As. Marcel Dekker, New York, USA. 2002. 363 – 380.
- Xiong, X., Liu, X., Iris, K.M., Wang, L., Zhou, J., Sun, X., Rinklebe, J., Shaheen, S.M., Ok, Y.S., Lin, Z. and Tsang, D.C. Potentially toxic elements in solid waste streams: Fate and management approaches. Environmental Pollution, 2019. 253, 680-707.
CrossRef - Ghosh, P., Rathinasabapathi, B., and Ma, L. Q. As-resistant bacteria solubilized As in the growth media and increased growth of As hyperaccumulator Pteris vittataL. Bioresource technology, 2011. 102(19), 8756-8761.
CrossRef - Farooq MA, Islam F, Ali B, Najeeb U,Mao B, Gill RA, Yan G, Siddique KHM and Zhou W As toxicity in plants: Cellular and molecular mechanisms of its transport and metabolism. Environmental and Experimental Botany. 2016. 132:42–52.
CrossRef - Laha A, Bhattacharyya S, Sengupta S, Bhattacharyya K, and GuhaRoy S, “Study on Burkholderiasp: Arsenic Resistant Bacteria Isolated from Contaminated Soil.” Applied Ecology and Environmental Sciences, 2021: 9(2). 144-148.
CrossRef - Titah, H. S., Abdullah, S. R. S., Idris, M., Anuar, N., Basri, H., Mukhlisin, M. and Kurniawan, S. B. Arsenic resistance and biosorption by isolated rhizobacteria from the roots of Ludwigiaoctovalvis. International journal of microbiology, 2018.
CrossRef - Li, K. and Ramakrishna, W. Effect of multiple metal resistant bacteria from contaminated lake sediments on metal accumulation and plant growth. Journal ofhazardous materials, 2011. 189(1-2), 531-539.
CrossRef - Srivastava, S., Verma, P.C., Chaudhry, V., Singh, N., Abhilash, P.C., Kumar, K.V., Sharma, N. and Singh, N. Influence of inoculation of As-resistant Staphylococcus arlettae on growth and As uptake in Brassica juncea (L.) Czern. Var. R-46. Journal of hazardous materials, 2013. 262, 1039-1047.
CrossRef - Carrasco, J. A., Armario, P., Pajuelo, E., Burgos, A., Caviedes, M. A., López, R., &Palomares, A. J. Isolation and characterisation of symbiotically effective Rhizobium resistant to arsenic and heavy metals after the toxic spill at the Aznalcollar pyrite mine. Soil Biology and Biochemistry, 2005. 37(6), 1131-1140.
CrossRef - Ma, Y., Prasad, M.N.V., Rajkumar, M. and Freitas, H. Plant growth promoting rhizobacteria and endophytes accelerate phytoremediation of metalliferous soils. Biotechnology advances, 2011. 29(2), 248-258.
CrossRef - Rajkumar, M., Sandhya, S., Prasad, M. N. V., and Freitas, H. Perspectives of plant-associated microbes in heavy metal phytoremediation. Biotechnology advances, 2012. 30(6), 1562-1574.
CrossRef - de-Bashan, L.E., Hernandez, J.P. and Bashan, Y. The potential contribution of plant growth-promoting bacteria to reduce environmental degradation– A comprehensive evaluation. Applied Soil Ecology, 2012. 61, 171-189.
CrossRef - Ghosh, P.K., Maiti, T.K., Pramanik, K., Ghosh, S.K., Mitra, S. and De, T.K. The role of As resistant Bacillus aryabhattai MCC3374 in promotion of rice seedlings growth and alleviation of As phytotoxicity. Chemosphere, 2018. 211, 407-419.
CrossRef - El-Meihy, R.M., Abou-Aly, H.E., Youssef, A.M., Tewfike, T.A., and El-Alkshar, E.A. Efficiency of heavy metals-tolerant plant growth promoting bacteria for alleviating heavy metals toxicity on sorghum. Environmental and Experimental Botany, 2019. 162, 295-301.
CrossRef - Sarkar, S., Basu, B., Kundu, C.K. and Patra, P.K. Deficit irrigation: An option to mitigate As load of rice grain in West Bengal, India. Agriculture, ecosystems & environment, 2012. 146(1), 147-152.
CrossRef - Sparks D.L., Page A.L., Helmke P.A., Leoppert R.H., Solthanpour P.N., Tabatabai M.A., Johnston C.T. and Sumner M.E. Methods of Soil Analysis. Part 3. Chemical Methods. Madison, Soil Science Society of America, 2006. 811–831.
- Johnston, S.E. and Barnard, W.M. Comparative Effectiveness of Fourteen Solutions for Extracting As from Four Western New York Soils 1. Soil Science Society of America Journal, 1979. 43(2), 304-308.
CrossRef - Kinegam, S., Yingprasertchai, T., Tanasupawat, S., Leepipatpiboon, N., Akaracharanya, A. and Kim, K.W. Isolation and characterization of As(III)-oxidizing bacteria from As-contaminated soils in Thailand. World Journal of Microbiology and Biotechnology, 2008.24(12), 3091-3096.
CrossRef - Hamza, T.A. and Alebejo, A.L. Isolation and Characterization of Rhizobia from Rhizospher and Root Nodule of Cowpea, Elephant and Lab LabPlants. International Journal of Novel Research in Interdisciplinary Studies, 2017. 4(4), pp.1-7.
- Majumder, A., Bhattacharyya, K., Bhattacharyya, S. and Kole, S.C. As-tolerant, As(III)-oxidising bacterial strains in the contaminated soils of West Bengal, India. Science of the total environment, 2013a.463, 1006-1014.
CrossRef - Majumder, A., Bhattacharyya, K., Kole, S. C., and Ghosh, S. Efficacy of indigenous soil microbes in arsenic mitigation from contaminated alluvial soil of India. Environmental Science and Pollution Research, 2013b.20(8), 5645-5653.
CrossRef - Holtz, J.D. Bergey's manual of determinative bacteriology. 1993. 9th ed. Baltimore: Williams and Wilkins.
- Dey, U., Chatterjee, S., and Mondal, N. K. Isolation and characterization of As-resistant bacteria and possible application in bioremediation. Biotechnology reports, 2016. 10, 1-7.
CrossRef - Cummings, D.E., Caccavo Jr., F., Fendorf, S., Rosenzweig, R.F. Arsenic mobilization by the dissimilatory Fe(III)-reducing bacterium Shewanella alga BrY. Environ. Sci. Technol. 1999. 33, 723-729.
CrossRef - Borch, T., Kretzschmar, R., Kappler, A., Cappellen, P.V., Ginder, V.A., Voegelin, A., Campbell, K. Biogeochemical redox processes and their impact on contaminant dynamics. Environ. Sci. Technol. 2010. 44, 15-23.
CrossRef - Sundararao, W.V.B. Phosphate dissolving organisms in the soil and rhizosphere. Indian. Jour. of Agr. Sci., 1963. 33, 272-278.
- Zaidi, S., Usmani, S., Singh, B.R. and Musarrat, J. Significance of Bacillus subtilis strain SJ-101 as a bioinoculant for concurrent plant growth promotion and nickel accumulation in Brassica juncea. Chemosphere, 2006. 64(6), 991-997.
CrossRef - Das, S., Jean, J.S., Kar, S., Chou, M.L. and Chen, C.Y. Screening of plant growth-promoting traits in As-resistant bacteria isolated from agricultural soil and their potential implication for As bioremediation. Journal of hazardous materials, 2014. 272, 112-120.
CrossRef - Penrose, D.M. and Glick, B.R. Methods for isolating and characterizing ACC deaminase containing plant growthâ€promoting rhizobacteria. Physiologia plantarum, 2003. 118(1), 10-15.
CrossRef - Reed, M.L.E. and Glick, B.R. Applications of plant growth-promoting bacteria for plant and soil systems. Applications of Microbial Engineering. Taylor and Francis, Enfield, CT, 2013. 181-229.
- Deepika, K.V., Raghuram, M., Kariali, E. and Bramhachari, P.V. Biological responses of symbiotic Rhizobium radiobacter strain VBCK1062 to the arsenic contaminated rhizosphere soils of mung bean. Ecotoxicology and environmental safety, 2016. 134, 1-10.
CrossRef - Liao, V.H.C., Chu, Y.J., Su, Y.C., Hsiao, S.Y., Wei, C.C., Liu, C.W., Liao, C.M., Shen, W.C. and Chang, F.J. As(III)-oxidizing and As(V)-reducing bacteria associated with As-rich groundwater in Taiwan. Journal of contaminant hydrology, 2011. 123(1-2), 20-29.
CrossRef - Botes, E., Van Heerden, E. and Litthauer, D. Hyper-resistance to As in bacteria isolated from an antimony mine in South Africa. South African Journal of Science, 2007. 103(7-8), 279-281.
- Titah, H.S., Abdullah, S.R.S., Anuar, N., Idris, M., Basri, H. and Mukhlisin, M. Isolation and screening of arsenic resistant rhizobacteria of Ludwigiaoctovalvis. African Journal of Biotechnology, 2011. 10(81), pp.18695-18703.
CrossRef - Yoon, I.H., Chang, J.S., Lee, J.H. and Kim, K.W. As(III) oxidation by Alcaligenes sp. strain RS-19 isolated from As-contaminated mines in the Republic of Korea. Environmental geochemistry and health, 2009. 31(1), 109.
CrossRef - Bachate, S.P., Khapare, R.M. and Kodam, K.M. Oxidation of As(III) by two β-proteobacteria isolated from soil., Applied microbiology and biotechnology, 2012. 93(5), 2135-2145.
CrossRef - Zhang, J., Xu, Y., Cao, T., Chen, J., Rosen, B. P., & Zhao, F. J. Arsenic methylation by a genetically engineered Rhizobium-legume symbiont. Plant and soil, 2017 416(1-2), 259-269.
CrossRef - Reichman, S.M. Probing the plant growth-promoting and heavy metal tolerance characteristics of Bradyrhizobium japonicum CB1809. European journal of soil biology, 2014. 63, 7-13.
CrossRef - Cavalca, L., Zanchi, R., Corsini, A., Colombo, M., Romagnoli, C., Canzi, E. and Andreoni, V. As-resistant bacteria associated with roots of the wild Cirsium arvense (L.) plant from an As polluted soil, and screening of potential plant growth-promoting characteristics. Systematic and applied microbiology, 2010. 33(3), 154-164.
CrossRef






