Histochemical Analysis of Expression Pattern of Camv 35S Promoter in Transgenic Rapeseed (Brassica Napus L.)
Parissa Jonoubi1 * , Ali Hatef Salmanian2 and Amin Ravaei1
1
Department of Plant Biology, Faculty of Biological Sciences,
Kharazmi University,
Tehran,
Iran
2
Department of Plant Biotechnology,
National Institute of Genetic Engineering and Biotechnology (NIGEB),
Tehran,
Iran
DOI: http://dx.doi.org/10.12944/CWE.10.Special-Issue1.90
Promoters are considered valuable tools for biotechnology and provide great opportunities for eugenic purposes. This study aimed to investigate the expression pattern of GUS gene directed by CaMV 35S promoter in transgenic rapeseed (Brassica napus L.). The GUS gene was transferred to the rapeseed explants by Agrobacterium. The regenerated and rooted transgenic plantlets were transferred to the pots containing a mixture of soli and vermiculite. These plants underwent PCR and histochemical GUS assay. The cross-section of leaf, petiole, and stem of the transformed plants was obtained. GUS activity was observed in phloem, parenchyma, collenchyma, and supporting tissue of vascular bundles. It was also observed in cambium, endoderm, vascular ray, pith parenchyma, epidermis, and trichomes. These results showed that CaMV 35S causes the expression of transgene in various tissues differently.
Copy the following to cite this article:
Jonoubi P, Salmanian A. H, Ravaei A. Histochemical Analysis of Expression Pattern of Camv 35S Promoter in Transgenic Rapeseed (Brassica Napus L.). Special Issue of Curr World Environ 2015;10(Special Issue May 2015). DOI:http://dx.doi.org/10.12944/CWE.10.Special-Issue1.90
Copy the following to cite this URL:
Jonoubi P, Salmanian A. H, Ravaei A. Histochemical Analysis of Expression Pattern of Camv 35S Promoter in Transgenic Rapeseed (Brassica Napus L.). Special Issue of Curr World Environ 2015;10(Special Issue May 2015). Available from: http://cwejournal.org?p=680/
Download article (pdf) Citation Manager Publish History
Select type of program for download
| Endnote EndNote format (Mac & Win) | |
| Reference Manager Ris format (Win only) | |
| Procite Ris format (Win only) | |
| Medlars Format | |
| RefWorks Format RefWorks format (Mac & Win) | |
| BibTex Format BibTex format (Mac & Win) |
Article Publishing History
| Received: | 2015-01-16 |
|---|---|
| Accepted: | 2015-02-11 |
Introduction
The current approach for foreign gene expression and recombinant protein production changes from prokaryotic systems toward plant systems due to the advantages of plants, which include high product quality, very low cost, high scale-up capacity, minor differences of glycosylation, low risk contamination, relatively low ethical concern, and inexpensive storage cost [(Ma et al., 2003)- (Egelkrout et al., 2012)]. According to International Service for the Acquisition of Agri-biotech Application's (ISAAA) report "2012 marked an unprecedented 100-fold increase in biotech crop hectarage from 1.7 million hectares in 1996 to 170 million hectares in 2012 – this makes biotech crops the fastest adopted crop technology in recent history – the reason – they deliver benefits" (James et al., 2012). In order to genetically transform plants, there is a need for regulatory promoters upstream of the putative gene to determine the site, level, and timing of the foreign gene expression (Egelkrout et al., 2012); these promoters are generally divided into five groups: 1) Constitutive promoters which cause the highest amount of expression in approximately all cell types. The most widely applied constitutive promoter in plant biotechnology that is highly expressed in most of the plant cells is the cauliflower mosaic virus (CaMV) 35S promoter (Yoshida et al., 2000). 2) Inducible promoters which cause the activating or deactivating of foreign gene expression in the presence or absence of factors such as wounding or pathogen invasion, chemical or physical inducements, and preferentially at the developmental stage which has no interference with the life cycle of the transgenic plant (Corrado et al., 2009). 3,4) Tissue-specific and development-stage-specific promoters are those that direct the expression of the transgene within the specific tissue such as leaves, seeds, roots, etc or during a specific stage of plant development that, in some cases, increase the stability of the produced protein [(Ma et al., 2003)- (Egelkrout et al., 2012)]. 5) Synthetic promoters which are composed of the regulatory elements of natural promoters (Sharma et al., 2009).
The localization of recombinant proteins is a considerable aspect of plant transformation which may affect the preservation and purification procedures of proteins (Wilken et al., 2012). Considering the chosen promoter, the localization of the foreign protein will be predictable and the application of reporter genes will facilitate this procedure. Using GUS gene as a reporter, it is possible to evaluate the function of novel promoters so that an appropriate choice can be made for the acute control of gene expression. Besides, the availability of routine histochemical analysis will greatly facilitate the studies of gene expression pattern during plant development. In addition, such methods are able to screen transformed cells and tissues more rapidly and sensitively.
As a constitutive promoter, the CaMV 35S promoter should naturally direct the expression of foreign gene in approximately all cell and tissue types; however, there is some evidence which shows different gene expressions in particular cells and tissues of the plants that imply different expression patterns (Anuar et al., 2011).
This study aimed to determine the GUS activity within the different tissues of the transgenic rapeseed to reveal the CaMV 35S promoter-directed gene expression.
Material and Methods
The Agrobacterium tumefaciens strain LBA4404 harboring pBI121 binary vector was used for the plant transformation. The T-DNA region of the vector had NPT II and GUS genes which were under the control of CaMV 35S promoter.
Transformation of Cotyledonary Explants by Agrobacterium
The Brassica napus var. PF 7045-91 and SLM seeds, after surface sterilization, were germinated in the jars containing half-strength MS (Murashige et al., 1962) media and maintained in a 16/8 h photoperiod at 40-50 µEm-2s-1 light intensity and 25°C in a greenhouse. The Agrobacterium was cultured overnight in LB (Luria-Bertani) broth containing 50mgl-1 kanamycin under shaking (180 rpm) at 28°C. The Agrobacterium culture with OD650= 0.8 was centrifuged at 3000rpm for 15 min. The pellet was resuspended at the infection medium containng MS salts and 5% glucose at pH 5.2 and used for the transformation of the explants. The cotyledons (with 5-7 mm petiole) of 7 day-old seedlings and hypocotyls from 14 day-old seedling were carefully excited. Only the petioles of the cotyledons and hypocotyl pieces were inoculated with Agrobacterium suspension for 5 to 10s. All the explants were wiped using a filter paper, then embedded at the MS medium without any hormone at pH 5.2, and maintained in the greenhouse under dark conditions at 28°C. After the co-cultivation, the hypocotyl explants were transferred to the callus induction medium (CIM) (MS medium containing 1 mgl-1 2,4-D, 30 gl-1 sucrose, 7 gl-1 agar, 200 mgl-1 cefotaxime, and 15 mgl-1 kanamycin at pH5.8) and then transferred to the shoot induction medium (SIM) (MS medium containing 1.5 mgl-1 benzyl adenine, 30 gl-1 sucrose, 7 gl-1 agar, 200 mgl-1 cefotaxime, and 15 mgl-1kanamycin at pH5.8) and subcultured every two weeks at the same fresh media. The cotyledonary explants were transferred to the SIM medium directly after the co-cultivation. After 6 weeks, the green kanamycin-resistant shoots were excited from the cotyledonary explants and transferred to the shoot maturation medium (SMM) (MS medium containing MS salt, 20 gl-1 sucrose, 7 gl-1 agar, 15 mgl-1 kanamycin, and 200 mgl-1 cefotaxime at pH 5.8). After 2 weeks, the green shoots were transferred to the root induction medium (RIM) (MS medium containing 2 mgl-1 indole butyric acid, 20 gl-1 sucrose, 6 gl-1 agar, 200 mgl-1 cefotaxime, and 15 mgl-1 kanamycin at pH 5.8). The kanamycin-resistance green plants which were able to produce roots, after developing the roots, were transferred to vermiculite, vermiculite and soil mixture, and soil, respectively.
PCR Confirmation of GUS Gene in Transgenic Plants
Genomic DNA of kanamycin-resistance plants was extracted according to the CTAB method (Murray et al., 1980) in order to be used for the presence of GUS gene in transgenic plants. Specific primers including GUS +2 and GUS -4 were exploited. The primer sequences were as follows:
GUS-4: 5'-CCGGCATAGTTAAAGAAATCATG-3'
GUS+2: 5'-GGTGGTCAGTCCCTTATGTTACG-3'
The amplification was done by TECHNE termocycler as follows: 30 cycles of denaturation at 94°C for 1 min, annealing at 61°C for 1 min, primer extension at 72°C for 1 min, and then final extension for 7 min at 72°C and initial denaturation at 94°C for 4 min.
GUS Assay
Evaluation of the GUS gene expression and production of β-glucoronidase enzyme, which produce the blue color by decomposition of X-Gluc, was conducted in the buffer containing50 mM phosphate buffer (pH 7), 1 mM X-GLUC, 1 mM EDTA, 0.001% triton, and 10 mM β-mercaptoethanol. The stem, petiole, and leaf pieces of the transgenic plants were incubated overnight at 37 °C in the buffer. For observing the blue color in the plant tissues, the elimination of chlorophyll was performed by 96% ethanol.
Transection Analysis
The vegetative tissues of the transgenic plants were analyzed by manual transverse section. The obtained sections were stained by carmine-vest and observed using a light microscope.
Results and Discussion
Using Agrobacterium-mediated transformation, the transformed plants from PF7045-91 and SLM cultivars with 10.2 and 12% transformation percentage were respectively obtained. Figure 1 shows the transformed and untransformed plants. Untransformed plantlets became white/violet at the selective media; however, the transformed plantlets maintained green with normal phenotype.
 |
Figure1: Effect of kanamycin as a selective agent on cotyledonary explants a) The transformed plantlets remain green in the presence of kanamycin; b) The untransformed plantlets became white/violet in the presence of kanamycin. Click here to View figure |
PCR confirmation of the GUS gene showed the presence of the gene on all of the putative transgenic lines. Figure 2 shows the 520 bp of PCR products indicating GUS gene. This result implied that the T-DNA region of the binary vector was successfully transferred to the plant cells.
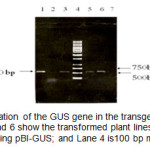 |
Figure2: PCR amplification of the GUS gene in the transgenic plants with specific primers. Lanes 3, 5, and 6 show the transformed plant lines; Lane 1 is the positive control using pBI-GUS; and Lane 4 is100 bp mix ladder. Click here to View figure |
Appearance of blue color in the tissues of transgenic plants indicated the expression of the GUS gene and function of the β-glucuronidase enzyme. In 73% of kanamycin-resistance plants, the blue color appeared at different intensities (Figure 3). In cross-section of the leaf, GUS activity was so high in vascular bundles that, in addition to phloem, vascular ray and pith parenchyma extremely displayed blue color. There was also different staining in the parenchymal cells of cortex and pith and also in epidermal cells, including the trichomes.
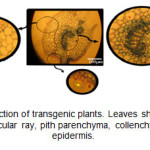 |
Figure3: Leaf cross-section of transgenic plants. Leaves show GUS expression in vascular bundle, vascular ray, pith parenchyma, collenchyma, trichomes, and epidermis. Click here to View figure |
In the petiole cross-section of the transgenic plants, the blue color was observed in phloem and collenchyma tissues (Figure 4).
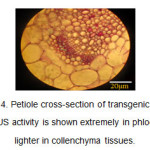 |
Figure4: Petiole cross-section of transgenic plants. The GUS activity is shown extremely in phloem and lighter in collenchyma tissues. Click here to View figure |
In the stem cross-section, the expression of GUS gene and appearance of blue color resulting from the decomposition of X-Gluc was highly observed in phloem tissues and vascular ray (Figure 5). GUS activity was highly seen in the phloem tissues around the vascular ring of the stem of CaMV-GUS plants (Jefferson et al., 1987). The expression pattern was punctuate and observable in the phloem parenchyma joining internal and external phloem (Esau K et al., 1977). In the stems of some transformed plants, variable distribution of GUS activity was observed.
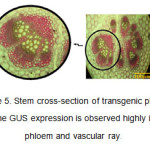 |
Figure5: Stem cross-section of transgenic plants. The GUS expression is observed highly in phloem and vascular ray. Click here to View figure |
It is absolutely clear that the patterns of GUS expression were intricate in the plant cells (Basu et al., 2004). Different metabolic activities were expected to be observed in different cell-types of plants; therefore, the GUS gene expression was affected by different biochemical, molecular, and biological factors (Fior et al., 2009), exhibited in rates of transcription and translation; such differences may be implied by the present findings. Alternatively, due to very small cross-sectional areas of phloem cells, the observed intense blue color may refer to greater cell number per unit area (Jefferson et al., 1987). During the S phase of cell cycle, the CaMV 35S promoter was so preferentially active that such an interpretation with the observed GUS expression pattern may imply the cell division within these tissues (Nagata et al., 1987). This remark originates from the 35S transcript role of CaMV in viral replication; some other plant DNA viruses like geminiviruses have such a replication in phloem tissues [ (Schubert et al., 2004) and (Dinant et al., 2004)]. Furthermore, CaMV 35S promoter has two domains: domain A contributes to meristematic tissue expression and domain B results in vascular bundle expression (Anuar et al., 2011). Considering the low or no endogenous GUS activity in higher plants, the observed localization may reflect a real difference in the expression level of CaMV 35S promoter in different cells.
Using Agrobacterium-mediated transformation, the transgenic cells and plants with different copies of foreign gene and also different sites of integration will be obtained. Thus, the transformed plants would express the foreign gene with different amounts. This difference may result from the varied localization of gene integration, influences of endogenous protein factors, and silencing during transcription or post-transcription [ (Wakimoto et al., 1998), 19, and (Schubert et al., 2004)]. High rate of putative gene insertion could result in the unstable or decreased level of gene expression. Transformation by Agrobacterium will result in a low rate of foreign gene insertion and DNA integration will be random with a tendency to be inserted in the distal chromosomal regions of the genome [(Dong et al., 2001) and (Gelvin, 2003)].
Now, great insight has been gained into transgenic plants and their features for the production of foreign proteins. Without the presence of a proper promoter, the foreign gene will be replicated, but not expressed; therefore, using appropriate promoters will result in the successful gene expression. Although the CaMV 35S promoter is a powerful promoter, it should not be taken into account as 'constitutive' anymore considering cell type- and cell cycle-based transcriptional processes. The availability of a great number of promoters makes the future very promising in terms of using plants as factories for the production of foreign proteins. Nowadays, the current knowledge about plant transformation and expression of foreign genes is in a good state and further works could be even more enlightening.
Acknowledgements
We thank the staffs of plant developmental biology laboratory of Kharazmi University and plant biotechnology laboratory of NIGEB for all assistances. The Study was supported by Kharazmi University and National Institute of Genetic Engineering and Biotechnology (NIGEB).
References
- Ma K.J. Drake M.P. Christou P., Nat. Rev. Genet., 4, 794 (2003).
- Sharma A.K. and Sharma M.K., Biotech. Adv., 27, 811 (2009).
- Egelkrout E. Rajan V. Howard J.A., Plant. Sci., 184, 83 (2012).
- James C., ISAAA Brief, (2012).
- Yoshida K. Shinmyo A., J. Biosci. Bioeng., 90, 353 (2000).
- Corrado G. Karali M., Biotech. Adv., 27, 733 (2009).
- Wilken L. Nikolov Z., Downstream processing of transgenic plant systems: Protein recovery and purification strategies. Molecular Farming in Plants: Recent Advances and Future Prospects, Springer, Netherlands, (2012).
- Anuar M.R. Ismail I. Zainal Z., J. Biotechnol., 10, 8236 (2011).
- Murashige T. Skoog F., Physiol. Plantarum., 15, 473 (1962).
- Murray M.G. Thompson W.F., Nucleic. Acids. Res., 8, 4321 (1980).
- Jefferson R.A. Kavanagh T.A. Bevan M.W., EMBO. J., 6, 3901 (1987).
- Esau K., The Anatomy of seed plants, Wiley, USA, (1977).
- Basu C. Kausch A.P. Chandlee J.M., Biochem. Biophys. Res. Commun., 320, 7 (2004).
- Fior S. Gerola P.D., Plant. Methods., 5, 1 (2009).
- Nagata T. Okada K. Kawazu T. Takebe I., MGG., 207, 242 (1987).
- Schubert D. Lechtenberg B. Forsbach A. Gils M. Bahadur S. Schmidt R., Plant. Cell., 16, 2561 (2004).
- Dinant S. Ripoll C. Pieper M. David C., Physiol. Plantarum., 121, 108 (2004).
- Wakimoto B.T., Cell., 93, 321 (1998).
- Wanger E.J. Garcia-Blanco M.A., Mol. Cell. Biol., 21, 3281 (2001).
- Dong J.J. Kharb P. Teng W.M. Hall T.C., Mol. Breeding., 7, 187 (2001).
- Gelvin S.B., MMBR., 67, 16 (2003).







