Benthic Foraminifera and Diatom Relationship: Insights from RbcL Gene Sequences in Palk Bay
1
Department of Marine Science,
Bharathidasan University,
Tiruchirappalli,
Tamil Nadu
India
Corresponding author Email: yoganandan@bdu.ac.in
DOI: http://dx.doi.org/10.12944/CWE.19.3.22
Copy the following to cite this article:
Balasubramaniyan M, Veeran Y. Benthic Foraminifera and Diatom Relationship: Insights from RbcL Gene Sequences in Palk Bay. Curr World Environ 2024;19(3). DOI:http://dx.doi.org/10.12944/CWE.19.3.22
Copy the following to cite this URL:
Balasubramaniyan M, Veeran Y. Benthic Foraminifera and Diatom Relationship: Insights from RbcL Gene Sequences in Palk Bay. Curr World Environ 2024;19(3).
Download article (pdf)
Citation Manager
Publish History
Select type of program for download
| Endnote EndNote format (Mac & Win) | |
| Reference Manager Ris format (Win only) | |
| Procite Ris format (Win only) | |
| Medlars Format | |
| RefWorks Format RefWorks format (Mac & Win) | |
| BibTex Format BibTex format (Mac & Win) |
Article Publishing History
| Received: | 2024-07-30 |
|---|---|
| Accepted: | 2024-11-01 |
| Reviewed by: | 
 Chadetrik Rout
Chadetrik Rout
|
| Second Review by: |

 Prachi Hatkar
Prachi Hatkar
|
| Final Approval by: | Dr. Gangadhar Andaluri |
Introduction
The shells of foraminifera, single-celled marine organisms with calcareous shells, are widely distributed and sensitive to environmental changes.1 Due to this sensitivity, variations in foraminiferal population and species assemblages are widely used as proxies for paleoclimatic and palaeoceanographic reconstructions.2 Several biological factors, including photosynthesis by symbionts, have been reported to contribute to the large variation observed in the disequilibrium of ?18O values.3, 4 The diversity of foraminifera and their symbiotic relationships with various groups of marine microorganisms have been extensively explored. 5,6 Studies have examined the planktonic foraminifera, benthic foraminifera, and large benthic foraminifera (LBF).7-10 Additionally, several researchers have investigated the cell wall morphology, mineral composition, and agglutination of foraminifera using scanning electron microscopy (SEM) and energy-dispersive X-ray (EDX) analysis.11-14 The mineral content has been analyzed to provide insight into their adaptations. Symbiosis, observed when two or more species of organisms interact closely, is known to confer mutual benefits, including nutrient exchange and protective mechanisms provided by the host.15 Certain species of foraminiferal have a symbiotic relationship with diatoms and microalgae, offering advantages to both partners. These symbiotic interactions have been demonstrated to influence the response of foraminifera to climate change, as the symbionts exhibit varied sensitivities to temperature, light, and pH.16 Through photosynthesis, energy and nutrients are obtained, which enhance growth, calcification rates, and survival in oligotrophic environments. Environmental stress, such as thermal stress, bleaching, and disease, is resisted by providing protection, antioxidants, or heat shock proteins and adapting to changing environmental conditions.17, 18 Therefore, understanding the role of symbiosis in foraminiferal ecology and evolution is essential for interpreting their fossil record and reconstructing past environmental changes.
The extraction of DNA from foraminifera was first attempted in studies,19–21 and the first rDNA sequence was reported in research.22 The LSU rDNA tree was confirmed by analyzing complete and partial sequencing of SSU rDNA sequences from benthic and planktonic foraminifera.23-25 DNA barcoding, a technique that does not depend on the species morphological characteristics, can accurately identify species.26 DNA barcoding employs short, standardised gene regions as internal species tags to enable rapid, precise, and automated species identification.27 Additionally, this molecular technique can facilitate diatom identification and diversity assessment using standardized genetic markers. Research on the foraminiferal barcode database includes not only DNA sequences but also taxonomic references, visual documentation, and metadata associated with the sequenced species.28-29 Initially, the mitochondrial Cytochrome c oxidase subunit 1 (CO1) gene was identified as a key marker for foraminifera. These foraminiferal CO1 give a better understanding of mitochondrial diversity and species morphology.30
Plant DNA barcoding studies examine chloroplast genome sequence variation in both coding and noncoding regions.31 The RbcL gene is widely used as a molecular marker in phylogenetic and taxonomic studies of diatoms and other photosynthetic organisms. Diatoms are important primary producers in aquatic ecosystems and serve as bioindicators of water quality. However, their identification based on morphology is often challenging and time-consuming. Moreover, the phylogenetic relationships among closely related diatom species may not be sufficiently resolved by the RbcL gene, as it exhibits a high homoplasy rate and a low variation level.32 This approach aims to enhance our understanding of foraminiferal taxonomy and has potential applications in areas such as diversity assessment, ecology, biogeography, and biomonitoring.26 This research found that important to study the biodiversity of both foraminifera and algae involved in these relationships and to understand whether certain species have specific partners.
The study aims to identify the symbiont relationship between algae in foraminifera. We present the first symbiont relationship RbcL gene sequence of Benthic foraminifera (Ammonia parkinsoniana or A.parkinsoniana) in Palk Bay.
Materials and Methods
Study Area
The Palk Bay region is located between Mandapam and Thondi in Tamil Nadu, India (Fig 1). This region, characterized by seagrasses colonization, supports a rich and diverse fauna, such as corals.33 Common foraminifera associated with the seagrass meadows include species from the genera Amphistegina, Rotaliidae, Spirillinidae, Trichohyalidae, Rosalinidae, and Elphidiidae.34 The mutualistic link between corals and their symbiotic algae permits coral reefs to exist. The region receives moderate freshwater input enriched with nutrients, enhancing productivity. During the summer, the maximum surface water temperature was 32°C, while the minimum was 27.5 ?.
 | Figure 1: A map depicting the study area located in Thondi, Palk Bay.
|
Table 1: Shows details about different sampling points in the study area
Site | Latitude | Longitude | Water Depth (Meters) |
S1 | 9?43’48’’N | 79?02’19’’E | 5 |
S2 | 9?42’54’’N | 79?04’46’’E | 4 |
S3 | 9?40’38’’N | 79?03’29’’E | 5 |
S4 | 9?38’06’’N | 79?01’57’’E | 5 |
S5 | 9?39’20’’N | 79?59’37’’E | 4 |
S6 | 9?41’53’’N | 79?01’11’’E | 6 |
Sample Collection
During the pre-summer monsoon and summer monsoon, we collected sediment samples at six different latitudes and longitude coordinates in Thondi, Palk Bay, Tamil Nadu, India (Fig 1; Table 1). Table 1 shows the sample location’s latitude, longitude, and water depth collected from Thondi. We used a grab sampler for the sediment collection, 50ml of wet sediment was taken from each location and was preserved in a 10% solution of neutralized formaldehyde for the foraminifera study. The surface water salinity and temperature at each station were measured using a reflectometer and a digital thermometer. After collection, the sediment was subsequently sieved through a 0.63 ?m mesh in tap water and treated with sodium hexametaphosphate. Living foraminifera and their distribution were examined under a stereo-zoom microscope.35, 36
An SEM was used to examine A. parkinsoniana images, magnified at a 15mm working distance with increasing magnifications.14 Additionally, EDX analysis was performed to assess the variations in elemental composition on the surface of the foraminifera. For molecular analysis, 100 mg of foraminiferal powder was homogenized using liquid nitrogen in a microcentrifuge tube. Table 2 lists the forward and reverse primers used in the present study. PCR amplification was carried out using a GeneAmp PCR System 9700 thermal cycler. Sequencing was performed with the BigDye Terminator v3.1 kit, using a GeneAmp PCR System 9700. The sequencing mix included distilled water, sequencing buffer, primers, and ExoSAP-treated PCR product. Sample were prepared with EDTA, sodium acetate, and ethanol, and sequenced using the ABI 3500 DNA Analyzer. Sequences quality was assessed with Sequence Scanner Software v1 (Applied Biosystems), and the sequences were aligned and edited using Geneious Pro v5.1.37
Table 2. Shows details of primers used for DNA barcoding studies from foraminifera.
Target | Primer Name | Direction | Sequence (5’ à 3’) |
Dt-RbcL | F56 | Forward | AGTGACCGTTACGAATCTGG |
R1010 | Reverse | AGGATCACCTTCTAATTTACC |
Result
Seasonal variation of primary parameter
The seasonal variation of A. parkinsoniana (Fig. 2) in the Thondi region was studied. Changes in A. parkinsoniana abundance with temperature and salinity across the two seasons (2023) are shown in Figure 3. During the pre-summer season, A. parkinsoniana abundance was higher in the region, measured at 88 individuals in station 1. Temperature is recorded at 28 °C, and the salinity level was 25 ppt (Fig. 3). In the summer season, benthic foraminifera (A. parkinsoniana) abundance in the region is measured at six individuals in station 4. The temperature was recorded at 30.1 °C, and the salinity level was 30 ppt (Fig. 3). During the pre-summer season, multiple regression analysis revealed a relationship between A. parkinsoniana and temperature with an R²=0.47, and between A. parkinsoniana and salinity with an R²=0.075. In the summer season, the analysis showed an R²=0.69 for the relationship between A. parkinsoniana and temperature, and an R²=0.30 for the relationship between A. parkinsoniana and salinity (Fig 4).
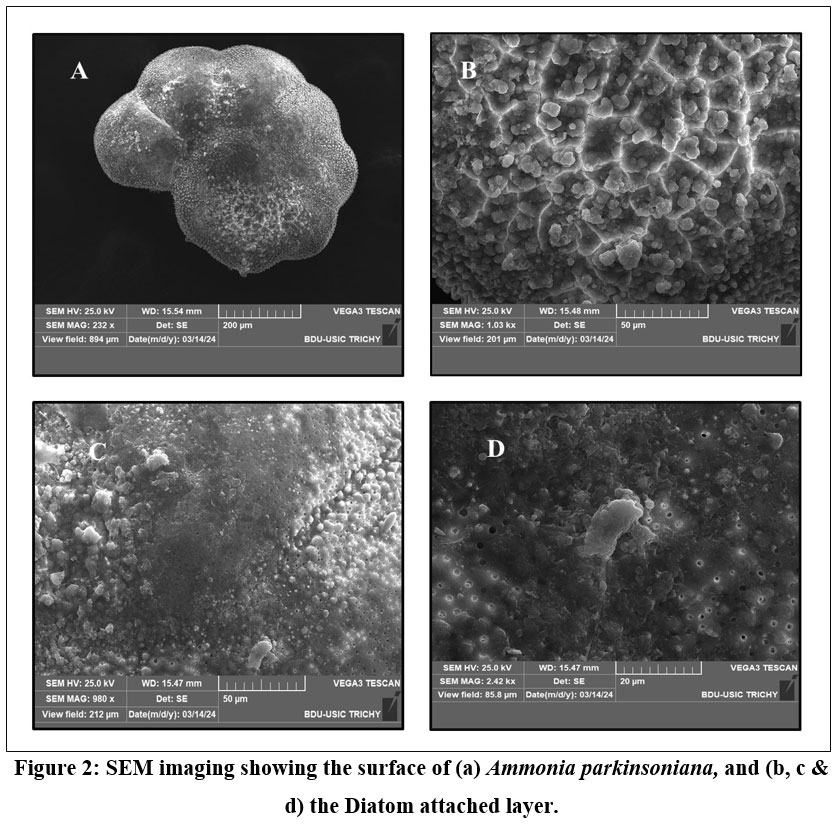 | Figure 2: SEM imaging showing the surface of (a) Ammonia parkinsoniana, and (b, c & d) the Diatom attached layer.
|
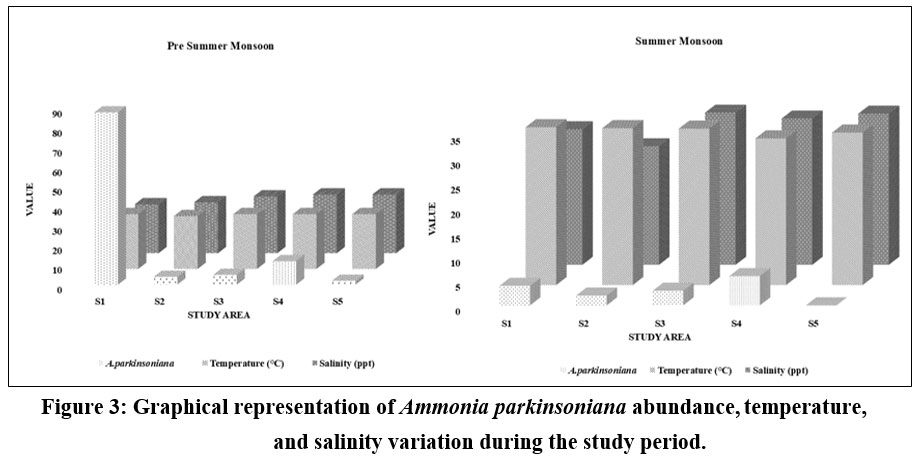 | Figure 3: Graphical representation of Ammonia parkinsoniana abundance, temperature, and salinity variation during the study period.
|
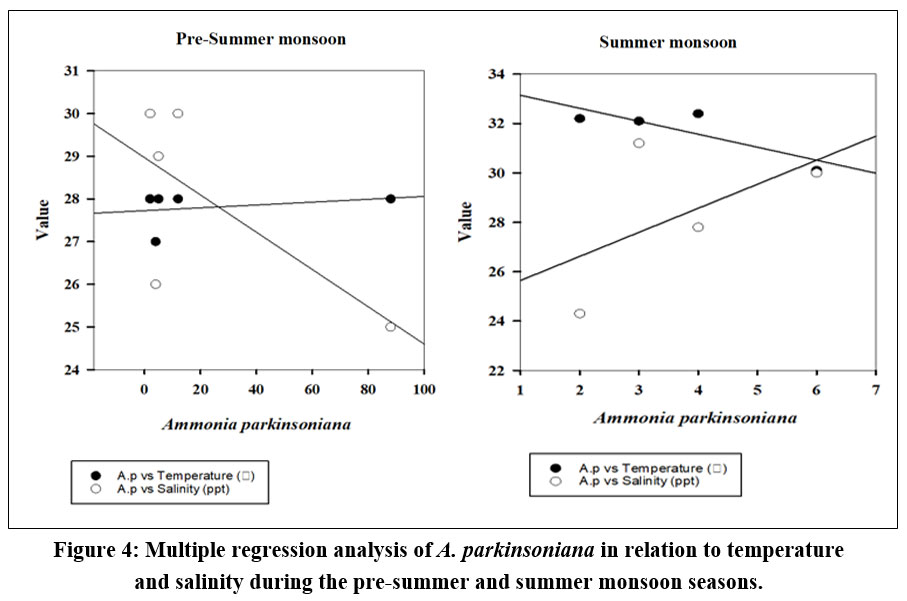 | Figure 4: Multiple regression analysis of A. parkinsoniana in relation to temperature and salinity during the pre-summer and summer monsoon seasons.
|
SEM EDX
SEM images of A. parkinsoniana revealed a clear indication of diatom species attachment on the surface (Fig 2). A tooth-like diatom attachment is shown in Figures 2c and 2d. Simultaneously, EDX analysis was used to detect significant changes in the elemental composition of foraminifera on both diatom-attached and non-attached surfaces. The EDX analysis of the non-attached area of A. parkinsoniana is illustrated in Figure 5, where a higher atomic percentage of carbon (35.34%) and an absence of silica were observed. The chamber walls of A. parkinsoniana specimens were found to be dominated by carbon (C), oxygen (O), and calcium (Ca). In the diatom-attached spot, a higher atomic percentage of oxygen (34.03%) and the presence of silica (1.46%) were detected. The diatom-identified area exhibited oxygen (O), calcium (Ca), carbon (C), and phosphorus (P). The findings suggest that diatoms significantly influence the elemental composition of A. parkinsoniana.
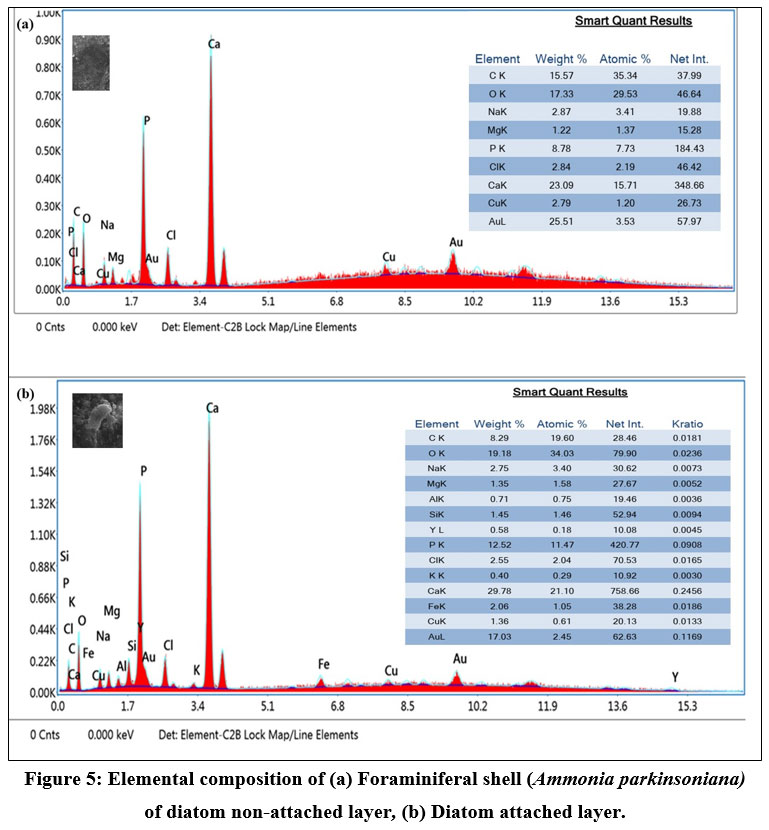 | Figure 5: Elemental composition of (a) Foraminiferal shell (Ammonia parkinsoniana) of diatom non-attached layer, (b) Diatom attached layer.
|
DNA Barcoding
The substantial genetic diversity among the specimens with diatom attachments was uncovered through additional DNA barcoding using the rcbL gene, subsequent to the preliminary SEM analysis of foraminifera. We isolated and amplified genomic DNA from A. parkinsoniana using a universal rbcL primer, and then sequenced the amplified DNA. We utilized BioEdit software version 5.0.6 to analyze the nucleotide composition of the rbcL gene sequence for Minutocellus sp. The sequence comprised 436 bp in the reverse direction and 700 bp in the forward direction, resulting in a total length of 1136 bp. Our result demonstrated that many diatom species taxa were consistent with the rbcL gene. The rbcL gene sequence of Minutocellu sp. was deposited in the GenBank of NCBI and assigned the accession number OR621302.
In this study, the rbcL gene of Minutocellu sp., a common marine planktonic diatom species that forms harmful algal blooms in coastal waters, was isolated and sequenced. The sequence was compared with other available rbcL sequences of Minutocellu sp. from different geographic regions, revealing high genetic variation within this species complex. Phylogenetic analysis was performed using rbcL sequences of other related diatom genera, and the evolutionary relationships were revealed. The results indicate that rbcL is an effective marker for diatom barcoding and phylogenetic studies, and suggest that Minutocellus sp. may represent a cryptic species complex, warranting further taxonomic revision.
Phylogenetic Tree
Phylogenetic trees are branch diagrams representing kinship relationships within a population or group.38 The phylogenetic tree was generated using MEGA 11.0 software, and various bootstrap analyses of the rbcL gene sequence were conducted employing both the Neighbor-Joining and UPGMA methods. A high similarity of 79% among the diatoms, with a neighbor-joining distance of 0.1 mm, was shown by the phylogenetic analysis (Fig. 6). A genetic distance of 25% between the foraminifera and diatom species is indicated by the scale height of 0.41 in the phylogenetic tree. The interspecific distance between all species was found to range from a higher distance scale of 0.41 to a lower distance scale of 0.001.
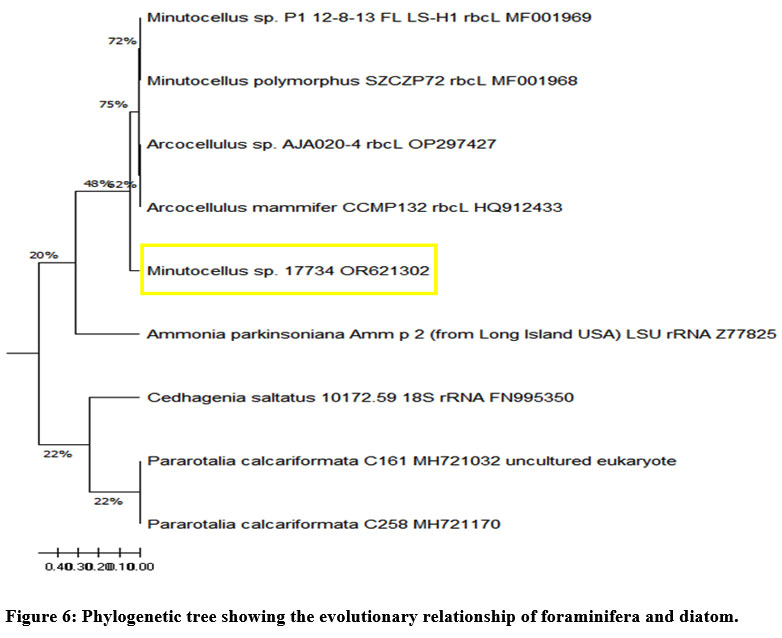 | Figure 6: Phylogenetic tree showing the evolutionary relationship of foraminifera and diatom.
|
Electropherogram Report
The electropherogram data report reveals that the highest peak corresponds to adenine (A) in the forward primer sequence. In contrast, the smallest peak is associated with guanine (G). Conversely, in the reverse primer sequence, the highest peak corresponds to guanine (G), with the smallest peak attributed to cytosine (C). Figure 7 indicates that adenine is the predominant base at a specific position in the DNA sequence, and guanine is the least frequent base at that location. Similarly, in the reverse primer, guanine is the most prevalent base, and cytosine is the least prevalent at that specific position in the sequence (Fig 8).
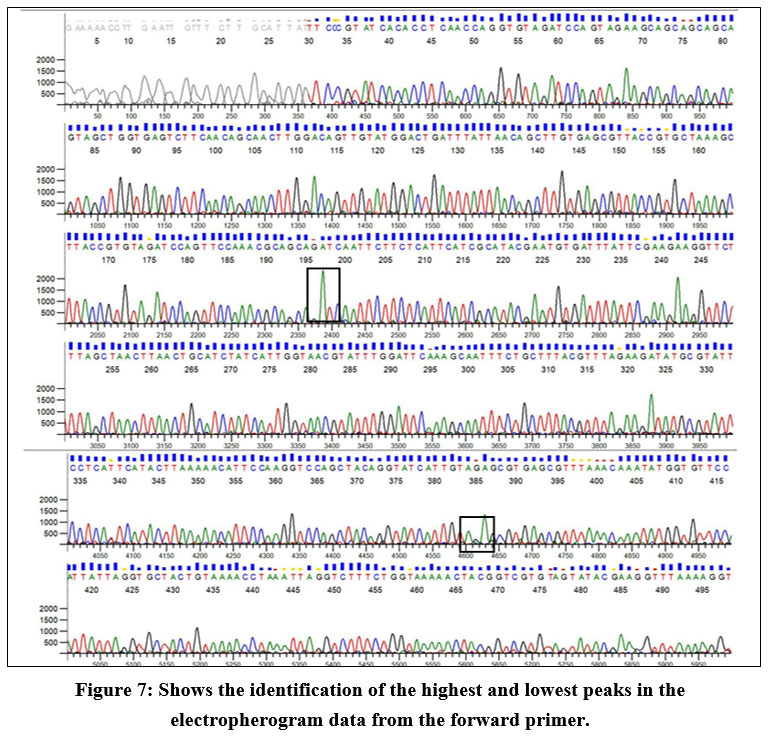 | Figure 7: Shows the identification of the highest and lowest peaks in the electropherogram data from the forward primer.
|
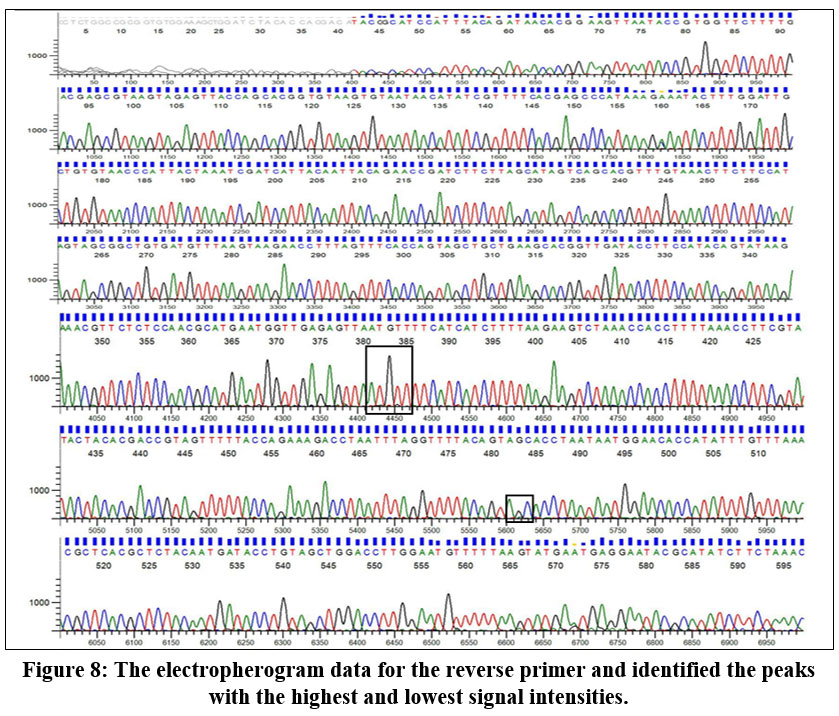 | Figure 8: The electropherogram data for the reverse primer and identified the peaks with the highest and lowest signal intensities.
|
Discussion
Our result confirms the presence of a symbiont relationship between algae and the foraminiferal shell. Using DNA barcoding to sequence genetically understudied taxonomic groups, we identified and developed chloroplast markers.
The present study demonstrates that (Table. 3) when temperature and salinity increase, the abundance of foraminifera decreases, indicating sensitivity to environmental changes. It is evident from these results 8 that local habitat plays a crucial role in shaping LBF's tolerance of changing environmental conditions; populations in stable environments show greater sensitivity to elevated temperatures and nitrate than populations in fluctuating environments, regardless of their fundamental tolerance ranges. Initial confirmation of the presence of diatom in foraminiferal studies was obtained using SEM analysis. This finding is consistent with the results of 13, who presented SEM images of fixed slides containing both foraminifera and diatoms. Additionally, they observed that individual foraminifera predominantly selected P. angulatum when introduced onto the slides as the dominant diatom species. Other studies revealed that the energy dispersive spectroscopy elemental analysis of Eggerella shells shows a dominated Ca:C molar ratio of 0.36.11 Additionally, we observed the same dominant element (Fig 8).
The symbiotic relationship between A. parkinsoniana benthic foraminifera and Minutocellu sp. diatoms holds promising implications for the survival and adaptation of these organisms in rapidly changing environments. A. parkinsoniana enhances an individual’s reactivity to environmental stimuli throughout all stages of development. Using biometric measurements along with qualitative traits enables a reliable classification of ammonia species.39 Two picoplanktonic diatoms were studied to examine how ecological niche adaptation relates to the ability and efficiency of photosynthetic regulation. One was found in the Pacific Ocean's upwelling zone (Minutocellus sp., strain RCC967) and the other in the Indian Ocean's open waters (Minutocellus sp., strain RCC703).40 The symbiotic relationship provides energy through the photosynthetic activity of diatoms and promotes calcification in foraminifera?.41 Our result shows benthic foraminifera and confirms the relationship with diatoms, consistent with some other results in the Israel coastal line.18 The diversity and flexibility of the algal symbiont community in globally distributed Amphistengina sp., a group of LBF widely used as bioindicators of environmental conditions.8 Various symbiont communities were found across geographical regions and environmental gradients, including rhodophytes, chlorophytes, diatoms, and dinoflagellates.42
Several studies have examined the relationship between foraminifera species and their symbionts.7-10, 18 Genotyping and symbiont cultures reveal that P. calcariformata hosts multiple diatom symbionts, which play a functional role throughout its life cycle, allowing the species to adjust its symbionts in response to stress.18 It was also suggested that changes in the types and quantities of algal symbionts over time in reaction to stress events could explain the unique thermal tolerance observed in P. calcariformata to date.43
The biology of symbiont-bearing foraminifera and discussed their potential as harbingers of global change, as well as the evolutionary history and biogeography of these associations.44 Many studies have investigated the symbiotic relationship between LBF and their algal partners, but few studies have performed experimental and cultured work on other benthic foraminifera species to confirm the symbiotic relationship in foraminifera. However, host adaptations and insights into the symbiosis in benthic environments. Foraminiferal symbionts are located just under the side walls, he concluded.7 In recent research, several diatom species have been found to associate with one individual of P. calcariformata, including species line Minutocellus polymorphus and Navicula sp.18 According to our study, the capacity to host various symbionts may enable shifts in symbiont community composition (symbiont shuffling), thereby enhancing foraminifera’s ecological success in dynamic environments. Additionally, specific dynamics between A. parkinsoniana and Minutocella sp. would require a full understanding of their unique interaction. This symbiotic association can lead to enhanced growth rates for the foraminifera due to the additional energy sources provided by the diatom. A. parkinsoniana provides habitat and carbon dioxide for Minutocella sp., which in turn, through photosynthesis, produces oxygen and organic compounds that the foraminifera can use.
The present study approach will enhance our comprehension of chloroplast diversity and evolution within these organisms facilitating the faster identification of species within this crucial yet insufficiently studied taxonomic group and other symbiont species. The symbiotic relationships will evolve or transform into new interactions if the environmental conditions change significantly and unpredictably. This may lead to new adaptations or innovations that enhance the survival or fitness of both foraminifera and diatom.
Conclusion
The results show the abundance of benthic foraminifera in both pre-summer and summer monsoon seasons, and how the abundance varies in each monsoon period in Palk Bay. Temperature significantly increased the abundance of the dominant A. parkinsoniana throughout the seasonal variations. A. parkinsoniana is commonly used as a bioindicator of environmental conditions, particularly in coastal and estuarine ecosystems. Furthermore, this study confirms the symbiotic relationship between benthic foraminifera (A. parkinsoniana) and diatoms (Minutocellus sp.). The symbiotic relationship between A. parkinsoniana and Minutocellus sp. holds promising implications for the survival and adaptation of these organisms in rapidly changing environments. This kind of study can help to understand the evolution of new adaptations and enhance the conservation of foraminifera and diatoms.
Acknowledgement
The author would like to thank Bharathidasan University, for granting the Ph.D. research work.
Funding Sources
The authors are thankful to Rashtriya Uchchattar Shiksha Abhiyan (RUSA 2.0.) for providing financial support (Project Grant no: 21-3/BDU/RUSA 2.0/TRP/BS/Date:08.10.2021).
Conflict of Interest
The author(s) do not have any conflict of interest.
Data Availability Statement
Data will be available depends upon the request.
Ethics Statement
This research did not involve human participants, animal subjects, or any material that requires ethical approval
Informed Consent Statement
This study did not involve human participants, and therefore, informed consent was not required
Author Contributions
Monisha Balasubramaniyan, Yoganandan Veeran conceived of the presented idea. Monisha Balasubramaniyan wrote the manuscript. Yoganandan Veeran verified the analytical methods. All authors discussed the results and contributed to the final manuscript.
References
- Van Dam, J. W., Negri, A. P., Mueller, J. F., Altenburger, R., & Uthicke, S. Additive pressures of elevated sea surface temperatures and herbicides on symbiont-bearing foraminifera. PLoS one. 2012; 7(3). 10.1371/journal.pone.0033900
CrossRef - Gooday, A.J. Benthic foraminifera (Protista) as tools in deep-water palaeoceanography: Environmental influences on faunal characteristics. Advances in Marine Biology. 2003; 46, 1-90.
CrossRef - Spero, H.J, Lea, D.W. Intraspecific stable isotope variability in the planktic foraminifera Globigerinoides sacculifer: results from laboratory experiments. Mar Micropaleontology. 1993; 22:221–234
CrossRef - Spero, H.J, Williams, D.F. Opening the carbon isotope “vital effect” black box 1. Seasonal temperatures in the euphotic zone. Paleoceanography. 1989; 4:593–601
CrossRef - Gastrich, M. D. Ultrastructure of a new intracellular symbiotic alga found within planktonic foraminifera 1. Journal of Phycology. 1987; 23(4), 623-632. https://doi.org/10.1111/j.1529-8817.1987.tb04215.x
CrossRef - Barnes, K. H. Diversity and distribution of diatom endosymbionts in Amphistegina spp. (Foraminifera) based on molecular and morphological techniques. 2016.
- Leutenegger, S. Symbiosis in benthic foraminifera - specificity and host adaptations. Journal of Foraminiferal Research. 1984; 14(1):16–35. https://doi.org/10.2113/gsjfr.14.1.16.
CrossRef - Prazeres M., Uthicke S., Pandolfi J.M. Influence of local habitat on the physiological responses of large benthic foraminifera to temperature and nutrient stress. Sci Rep. 2016; 6(1): 21936. https://doi.org/10.1038/srep21936.
CrossRef - Boudagher-Fadel MK. Evolution and geological significance of larger benthic foraminifera. Wignall PB, editor. Amsterdam: Elsevier. 2008.
- Lee J. J, Hallock P. Algal symbiosis as the driving force in the evolution of larger Foraminifera. Ann N Y Acad Sci. 1987; 503(1):330–47. https://doi.org/10.1111/j.1749-6632.1987.tb40619.x.
CrossRef - Bertram, M. A., & Cowen, J. P. Biomineralization in agglutinating foraminifera: an analytical SEM investigation of external wall composition in three small test forms. Aquatic Geochemistry. 1998; 4(3), 455-468. https://doi.org/10.1023/A:1009648701741
CrossRef - Grønlund, H. A. N. S., & Hansen, H. J. Scanning electron microscopy of some recent and fossil nodosariid foraminifera. Bulletin of the Geological Society of Denmark. 1976; 25(3–4), 121-134.
CrossRef - Austin, H. A., Austin, W. E., & Paterson, D. M. Extracellular cracking and content removal of the benthic diatom Pleurosigma angulatum (Quekett) by the benthic foraminifera Haynesina germanica (Ehrenberg). Marine Micropaleontology. 2005; 57(3-4), 68-73. https://doi.org/10.1016/j.marmicro.2005.07.002
CrossRef - Mancin, N., Basso, E., Kaminski, M. A., & Dogan, A. U. A standard SEM-EDS methodology to determine the test microstructure of fossil agglutinated foraminifera. Micropaleontology. 2014; 13-26. https://www.jstor.org/stable/24413636.
CrossRef - Caron D. Symbiosis and mixotrophy among pelagic microorganisms. In: Kirchman DL (ed) Microbial ecology of the oceans. Wiley-Liss, Inc., New York, 2000; pp 495–523
- Lee, J. J. Fueled by symbiosis, Foraminifera have evolved to be giant complex protists. All Flesh Is Grass: Plant-Animal Interrelationships, 2011; 427-452. https://doi.org/10.1007/978-90-481-9316-5_20
CrossRef - Prazeres, M., Roberts, T. E., Ramadhani, S. F., Doo, S. S., Schmidt, C., Stuhr, M., & Renema, W. Diversity and flexibility of algal symbiont community in globally distributed larger benthic foraminifera of the genus Amphistegina. BMC microbiology. 2021; 21(1), 1-16. https://doi.org/10.1186/s12866-021-02299-8
CrossRef - Schmidt, C., Morard, R., Romero, O., & Kucera, M. Diverse internal symbiont community in the endosymbiotic foraminifera Pararotalia calcariformata: implications for symbiont shuffling under thermal stress. Frontiers in Microbiology. 2018; 9. https://doi.org/10.3389/fmicb.2018.02018
CrossRef - Langer, M.R., Lipps, J.H. and Piller, W.E. Molecular paleobiology of protists: amplification and direct sequencing of foraminiferal DNA. Micropaleontology. 1993; 391. https://doi.org/10.2307/1485975.
CrossRef - Wray, C.G., Lee, J.J. and De Salle, R. Extraction and enzymatic characterization of foraminiferal DNA. Micropaleontology. 1993; 39: 69-73. https://doi.org/10.2307/1485976.
CrossRef - Stathoplos, L. and Tuross, N. Proteins and DNA from modem planktonic foraminifera. Journal of Foraminiferal Research. 1994; 24:49 -59.
CrossRef - Pawlowski, J., Bolivar, I., Guiard-Maffia, J. and Gouy, M. Phylogenetic position of foraminifera inferred from LSU rRNA gene sequences. Molecular Biology and Evolution. 1994; 11:929-938. https://doi.org/10.1093/oxfordjournals.molbev.a040174
CrossRef - Pawlowski, J., Bolivar, I., Fahrni, J. F., Cavalier-Smith, T., & Gouy, M. Early origin of foraminifera suggested by SSU rRNA gene sequences. Molecular Biology and Evolution. 1996; 13(3), 445-450. https://doi.org/10.1093/oxfordjournals.molbev.a025605
CrossRef - Darling, K. F., Kroon, D., Wade, C. M., & Leigh Brown, A. J. Molecular phylogeny of the planktic foraminifera. The Journal of Foraminiferal Research. 1996; 26(4), 324-330.
CrossRef - Wade, C. M., Darling, K. F., Kroon, D., & Leigh Brown, A. J. Early evolutionary origin of the planktic foraminifera inferred from small subunit rDNA sequence comparisons. Journal of molecular evolution. 1996; 43, 672-677. https://doi.org/10.1007/BF02202115
CrossRef - Paul, J. J. P., & Udhaya, C. I. DNA Barcoding and Molecular Taxonomy of Gracilaria Fergusonii J. Ag. Using rbcL Gene. World. 2020; 9(1), 49-53.
- Hebert, P. D., & Gregory, T. R. The promise of DNA barcoding for taxonomy. Systematic biology. 2005; 54(5), 852-859. DOI: 0.1080/10635150500354886.
CrossRef - Pawlowski, J., & Holzmann, M. A plea for DNA barcoding of foraminifera. The Journal of Foraminiferal Research. 2014; 44(1), 62-67. https://doi.org/10.2113/gsjfr.44.1.62.
CrossRef - Pawlowski, J. Introduction to the molecular systematics of foraminifera. Micropaleontology. 2000; 46, 1-12. https://archive-ouverte.unige.ch//unige:171296.
- Macher, J. N., Wideman, J. G., Girard, E. B., Langerak, A., Duijm, E., Jompa, J., ... & Renema, W. First report of mitochondrial COI in foraminifera and implications for DNA barcoding. Scientific reports. 2021; 11(1), 22165. https://doi.org/10.1038/s41598-021-01589-5.
CrossRef - Chen, S. L., Yao, H., Han, J. P., Liu, C., Song, J. Y., Shi, L. C., ... & Leon, C. Validation of the ITS2 region as a novel DNA barcode for identifying medicinal plant species. PLoSone. 2010; 5, e8613. https://doi.org/10.1371/journal.pone.0008613
CrossRef - Medlin, L., Jung, I., Bahulikar, R., Mendgen, K., Kroth, P., & Kooistra, W. H. C. F. Evolution of the diatoms. VI. Assessment of the new genera in the araphids using molecular data. Nova Hedwigia. 2008; 133, 81-100.
- Sivaleela, G., Samuel, D., Anbalagan, T., & Shrinivaasu, S. Seagrass associated marine sponges in Palk Bay. Records of the Zoological Survey of India. 2013; 113(3), 97-103.
CrossRef - Walton, W. R. Techniques for recognition of living foraminifera. Cushman Found. Foram. Res. Contr. 1952; 3(2), 56-60.
- Mateu-Vicens, G., Box, A., Deudero, S., & Rodri?guez, B. Comparative analysis of epiphytic foraminifera in sediments colonized by seagrass Posidonia oceanica and invasive macroalgae Caulerpa spp. The Journal of Foraminiferal Research. 2010; 40(2), 134-147. https://doi.org/10.2113/gsjfr.40.2.134
CrossRef - Hemleben, C., Spindler, M., Anderson, O. R., Hemleben, C., Spindler, M., & Anderson, O. R. Taxonomy and species features. Modern planktonic foraminifera. 1989; 8-32. 10.1007/978-1-4612-3544-6_2.
CrossRef - Drummond AJ, Ashton B, Buxton S, Cheung M, Cooper A, Heled J, Kearse M, Moir R, StonesHavas S, Sturrock S, Thierer T, Wilson A. (2010)– Geneious v. 5.1, Available from: http://www.geneious.com
- Nei, M., & Kumar, S. Molecular evolution and phylogenetics. Oxford University Press, USA. 2000.
CrossRef - Schönfeld, J., Beccari, V., Schmidt, S., & Spezzaferri, S. Biometry and taxonomy of adriatic Ammonia species from Bellaria–Igea Marina (Italy). Journal of Micropalaeontology. 2021. 40(2), 195-223. https://doi.org/10.5194/jm-40-195-2021
CrossRef - Giovagnetti, V., Cataldo, M. L., Conversano, F., & Brunet, C. Growth and photophysiological responses of two picoplanktonic Minutocellus species, strains RCC967 and RCC703 (Bacillariophyceae). European journal of phycology. 2012. 47(4), 408-420. https://doi.org/10.1080/09670262.2012.733030
CrossRef - Guillermic, M., Misra, S., Eagle, R., Villa, A., Chang, F., & Tripati, A. Seawater pH reconstruction using boron isotopes in multiple planktonic foraminifera species with different depth habitats and their potential to constrain pH and pCO2 gradients. Biogeosciences. 2020; 17(13), 3487-3510. https://doi.org/10.5194/bg-17-3487-2020
CrossRef - Xu, S., Chen, J., Qin, M., Jiang, L., & Qiao, G. Geography-dependent symbiont communities in two oligophagous aphid species. FEMS Microbiology Ecology. 2021; 97(10), fiab132. https://doi.org/10.1093/femsec/fiab132
CrossRef - Schmidt, C., Morard, R., Almogi-Labin, A., Weinmann, A. E., Titelboim, D., Abramovich, S., & Kucera, M. Recent invasion of the symbiont-bearing foraminifera Pararotalia into the Eastern Mediterranean facilitated by the ongoing warming trend. PLoS One. 2015. 10(8), e0132917. https://doi.org/10.1371/journal.pone.0132917
CrossRef - Lee, J. J., Cervasco, M. H., Morales, J., Billik, M., Fine, M., & Levy, O. Symbiosis drove cellular evolution: symbiosis fueled evolution of lineages of Foraminifera (eukaryotic cells) into exceptionally complex giant protists. Symbiosis. 2010; 51, 13-25. https://doi.org/10.1007/s13199-010-0056-4
CrossRef






