Biodegradation of Chlorpyrifos by Pseudomonas Resinovarans Strain AST2.2 Isolated from Enriched Cultures.
Anish Sharma 1 * , Jyotsana Pandit 1 , Ruchika Sharma 1 and Poonam Shirkot 1
Corresponding author Email: janish.sharma28@gmail.com
DOI: http://dx.doi.org/10.12944/CWE.11.1.33
A bacterial strain AST2.2 with chlorpyrifos degrading ability was isolated by enrichment technique from apple orchard soil with previous history of chlorpyrifos use. Based on the morphological, biochemical tests and 16S rRNA sequence analysis, AST2.2 strain was identified as Pseudomonas resinovarans. The strain AST2.2 utilized chlorpyrifos as the sole source of carbon and energy. This strain exhibited growth upto 400mg/l concentration of chlorpyrifos and exhibited high extracellular organophosphorus hydrolase (OPH) activity. Gas chromatography-flame ionization detector (GC-FID) studies revealed that Pseudomonas resinovarans AST2.2 degraded 43.90 % of chlorpyrifos (400 mg/l) within 96 hrs. Intermediates of chlorpyrifos degradation were identified using GC-MS. This strain have potential to degrade chlorpyrifos and thus can be used for bioremediation and ecological restoration of sites contaminated with chlorpyrifos.
Copy the following to cite this article:
Sharma A, Pandit J, Sharma R, Shirkot P. Biodegradation of Chlorpyrifos by Pseudomonas Resinovarans Strain AST2.2 Isolated from Enriched Cultures. Curr World Environ 2016;11(1) DOI:http://dx.doi.org/10.12944/CWE.11.1.33
Copy the following to cite this URL:
Sharma A, Pandit J, Sharma R, Shirkot P. Biodegradation of Chlorpyrifos by Pseudomonas Resinovarans Strain AST2.2 Isolated from Enriched Cultures. Curr World Environ 2016;11(1). Available from: http://www.cwejournal.org/?p=13906
Download article (pdf) Citation Manager
Select type of program for download
| Endnote EndNote format (Mac & Win) | |
| Reference Manager Ris format (Win only) | |
| Procite Ris format (Win only) | |
| Medlars Format | |
| RefWorks Format RefWorks format (Mac & Win) | |
| BibTex Format BibTex format (Mac & Win) |
Introduction
Organophosphorus pesticides are most commonly employed in insect pest management for crop production, municipal hygiene, and disease vector control.1 Chlorpyrifos [O,O-diethyl-O-(3,5,6-trichloro-2- pyridinyl)phosphorothioate] is extensively used broad-spectrum insecticide against gall midge, cutworms, corn root worms, leaf folder, leaf hopper, etc.2 Chlorpyrifos is accessible in variety of formulations like granules, wet table powder, dustable powder and emulsifiable concentrate.3
Comprehensive and voluminous usage and handling of chlorpyrifos contaminates air, ground water, water bodies and soil. Excessive pesticide usage resulted in accumulation of pesticide residues in crops, soils, and biosphere creating an ecological stress.4 Many reports from Environmental Protection Agency have been submitted regarding contamination of water and terrestrial ecosystems with chlorpyrifos.5 Researchers had reported that variation in factors, like pH, temperature, organic carbon content, moisture content, and pesticide formulation affects half-life of chlorpyrifos in soil and it varies from 10 to 120 days. 3,5,6-trichloro-2-pyridinol (TCP) is the major degradation product which has greater water solubility than chlorpyrifos and results in widespread contamination of soils and aquatic environment.6 Chlorpyrifos and its residues has been identified in marine sediments, agricultural soils, streams, rivers, urban storm drains, freshwater lakes, groundwater, fog, rain and air.7
Degradation of chlorpyrifos employing microbes is of particular concern because of its high mammalian toxicity and widespread usage in agricultural sector. Biodegradation is considered to be a reliable cost-effective technique for pesticides abatement and a major factor determining the fate of organophosphorus pesticides in the environment.8 Mallick et al., 19999 reported that Arthrobacter species degraded chlorpyrifos in mineral salt medium and this bacterium was initially isolated from methyl parathion-enriched soil. Employing enrichment procedure10 isolated six chlorpyrifos-degrading bacteria. Many chlorpyrifos -degrading bacteria including members of genera Arthrobacter sp.,11 Enterobacter strain B-14,12 Alcaligenes faecalis DSP3,13 Klebsiella sp.,14 Sphingomonas sp. Dsp-2,15 Pseudomonas aeruginosa NCIM 2074,16 Bacillus pumilus C2A1,17 Pseudomonas putida MAS-1,18 Cupriavidus sp.,19 had been isolated from environment contaminated with the pesticide.
The main objective of the present study involved the isolation and identification of chlorpyrifos-utilizing bacteria from apple orchard soil using an enrichment culture technique and degradation of chlorpyrifos by the bacterial isolate in liquid medium. The study aims at explicate a possible employment of an isolated bacterial strain for remediation of the chlorpyrifos-contaminated environment.
Material and Methods
Collection of Soil Sample
Soil sample were collected from apple orchard soil contaminated with various pesticides from Theog site, Shimla district, Himachal Pradesh, India, where chlorpyrifos had been used extensively to control various pests. Soil samples were collected from the top soil ranging in depth from 10-15cm using sterilized spatula.20 Samples were collected in triplicates and brought to laboratory for further experiments.
Chemicals and Media
Chlorpyrifos (O, O-diethyl O-(3, 5, 6-trichloro-2-pyridyl) phosphorothioate) (99.6 % pure analytical grade) was obtained from Sigma-Aldrich, Switzerland. Samples of methyl parathion (99.9 % purity) were obtained from Supleco Analytical, USA. Mineral salt medium (MSM) was employed for enrichment, isolation and characterization of chlorpyrifos degrading bacteria.10 The MSM comprised of (g/l) 1.5 g K2HPO4, 0.5 g KH2PO4, 0.5 g (NH4)2SO4, 0.5 g NaCl, 0.2 g MgSO4, 0.05 g CaCl2, 0.02 g FeSO4, 1 liter distilled water with pH 7.0. Sterilization was done by autoclaving media at 121°C and 15 psi for 15 minutes.
Enrichment Technique and Isolation
The soil samples were air dried at room temperature and moisture content was retained to nearly 20% (w/w). These soil samples were sieve through 2 mm mesh. About 10 g of soil sample were added in 250 ml Erlenmeyer flask containing 50 ml of mineral salt medium (MSM) with 10 mg/l concentration of chlorpyrifos. The samples were incubated in dark on a shaker incubator at 37°C and 150 rpm for one week. After one week of incubation 10 ml culture was transferred to flask with fresh MSM with 30 mg/l concentration of chlorpyrifos and incubated for another week. Further 1 ml of culture was transferred to fresh MSM with 50 mg/l chlorpyrifos and incubated at 37°C and 150 rpm for another week. Chlorpyrifos degrading as well as resistant bacteria pure cultures were then isolated by spread plate technique and streak plate method. 100 µl of enriched culture was surface plated on the agar solidified MSM fortified with 50 mg/l chlorpyrifos in glass petri plates and incubated at 37°C after 24 hours, bacterial colonies which appeared on the medium, were individually streak purified, and then maintained on MSM agar medium slants at 4°C and as glycerol stocks at -20°C.
Chlorpyrifos Resistance Profile of Bacterial Isolates
Each of the selected chlorpyrifos degrading bacterial isolates was streaked on mineral salt medium containing varying concentration of chlorpyrifos ranging from 50-800 mg/l.21 Bacterial isolates were streaked on MSM agar plates fortified with chlorpyrifos and incubated for 48 hours at 37°C. Bacterial isolate exhibiting growth at highest concentration of chlorpyrifos was then selected for further studies.
Characterization of Chlorpyrifos Degrading Bacterial Isolate
Purified bacterial isolate with tolerance to highest chlorpyrifos concentration was characterized on morphological characters and as well as employing biochemical tests like Methyl red & Voges-Proskauer (MR-VP), Catalase, Citrate utilization, Oxidase, Urease, Casin hydrolase, fermentation of glucose, sucrose and lactose. Bacterial isolate was taxonomically identified from Bergey’s manual of systematic bacteriology and further confirmation was made by sequencing 16S rRNA gene. Genomic DNA extraction kit-Mini (Real Genomics) was used for extraction of total genomic DNA of selected isolates. Nanodrop spectrophotometer (NanoDrop ND1000, Thermo Fisher Scientific, USA) was employed to quantify DNA. Amplification of 16S rRNA gene was done by PCR utilizing universal primers 27f (5- AGAGTTTGATCCTGGCTCAG-3, forward) and 1492r (5'-GGTTACCTTGTTACGACTT-3', reverse). Automated sequencer (Genetic analyzer 31030, Accessories Applied Biosystems) was used to carry out sequencing. Using the EzTaxon server, phylogenetic neighbours were identified and the pairwise 16S rRNA sequence similarities were calculated [22]. MEGA6 software package was utilized for Phylogenetic analysis, after multiple alignments of data available from public databases by CLUSTALW.23
Quantitative Screening for Chlorpyrifos Degrading OPH Activity
Selected bacterial isolate was further screened for organophosphate hydrolase (OPH) activity by quantitative method. Bacterial cells were cultured in 1.0 liter of Luria Bertani (LB) medium overnight. Overnight cultures of bacterial isolate was used to obtain culture supernatant at 8,000 x g, 4°C for 10 minutes and this was further used as crude extracellular enzyme. Wash cell pellets twice and then resuspend it in 50 mmol/l Tris-Cl (pH 8.0) buffer containing 0.1 mmol/l phenylmethylsulfonyl fluoride (PMSF). Cell disruption was done by sonication 10 times for 10 seconds period at 15 seconds intervals, by using a (Digital Sonifier-MP Biomedical). Unbroken cells were removed by centrifugation at 15000 × g for 30 minutes and cell free supernatant obtained was used as crude intracellular enzyme. Crude intracellular and extracellular enzyme preparations were used for further quantitative assays.24
Hundred μl of crude intracellular and extracellular enzyme preparations was added to 900 μl of 50 mmol/l Tris-Cl (pH 9.0) assay buffer incorporated with 5.0 μl of 10 mg/ml methyl-parathion as a substrate and incubated at 37°C for 10 minutes. Termination of the reaction was done by addition of 1.0 ml of 10 % CCl3COOH (Trichloroacetic acid) followed by addition of 1.0 ml of 10 % Na2CO3 for color development. The absorbance was recorded at 410 nm wavelength using spectrometer (Perkin Elmer Lambda 25). According to the amount of p-nitrophenol (PNP) the product of hydrolysis and enzyme activity was calculated. One unit of OPH activity is defined as the amount of enzyme liberating 1.0 μmol of p-nitrophenol per minute at 37°C.
Analytical Procedure
Preparation of Inoculum
Bacterial strain AST2.2 was cultured and raised to exponential phase using mineral salt medium, and then collected by centrifugation at 5000 g for 5 mins at room temperature. The cell pellets were washed twice with sterilized MSM and adjusted to approximately 3x108 CFU/ml for degradation experiments.
Inoculation and Fortification of Mineral Salt Media (MSM)
Hundred μl of bacterial culture suspension containing 3x108 CFU/ml was inoculated into flasks containing 20 ml minimal medium enriched with 400 mg/l of chlorpyrifos. Un-inoculated flasks with minimal medium (containing 400 mg/l chlorpyrifos) were used as control. The flasks were incubated at 37ËšC for different time of intervals (0, 24, 48, 72 and 96 hours).
Extraction and Partitioning
Liquid-liquid extraction method was performed for extraction of chlorpyrifos and its residues from the bacterial culture. Chlorpyrifos containing culture was transferred to separating funnel and 20 ml n-hexane was added to medium. The mixture was shaken vigorously for 4-5 minutes and was kept undisturbed, until separation of two liquids took place. The samples were extracted three times and n-hexane layer was collected in 250 ml conical flasks. The extract was evaporated on rotary evaporator at 50°C to almost dryness under pressure employing vacuum pump and residue was re-dissolved in 2.0-5.0 ml n-hexane in sterile glass vials for GC determination.
Chromatographic Determination
GC-FID analysis of various extracts was performed on a Shimadzu GC-17A gas chromatograph. The column size was 30 m x 0.25 mm internal diameter with Optima-5 (Macherey-Nagel) capillary column. The composition of the capillary column was 5 % phenyl-95 % methylpolysiloxane. The analysis was performed in the split mode with 1:10 ratio. The column and injector temperatures were maintained at 300°C and 400°C respectively and detector temperature was maintained at 450°C. The column pressure was 77 kPa with 1.0 ml/min column flow and the total flow was 12 ml. The carrier gas used was nitrogen.18 Percentage recovery of chlorpyrifos was calculated as follows:
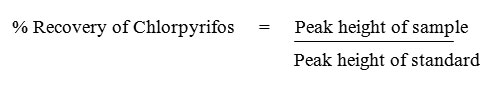
Amount of chlorpyrifos degraded = Original amount added – Amount recovered
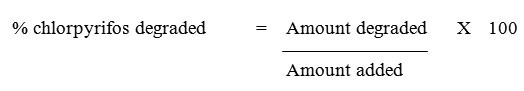
GC-MS for Determination of Chlorpyrifos Intermediates
GC-MS was performed with a TRACE 1300GC TSQ 8000 assembled with a split-splitless injector and connected to a triple quadrupole mass spectrometer. TG-5 MS capillary column (30 m × 0.25 mm, 0.25 μm, containing 5 % phenylmethylpolysiloxane) (J&W Scientific, Santa Clara, CA) was used for separation.25 Helium (99.99 %) was used as carrier gas at a constant flow rate of 1 ml/min. Injector temperature was kept at 250ºC in split less mode (5 minutes), and oven temperature was programmed as follows: initial temperature 100°C (hold 2 minutes), 20°C per min to 180°C, and 10°C per minute to 250°C (hold 2 minutes). The total GC–MS analysis time was 15 minutes. Electron ionization mode was used for MS ionization and spectra were recorded at 70 eV. The GC–MS interface and the ion source temperature were set at 250ºC and 200ºC, respectively. The total ion current (TIC) chromatograms were recorded between 40-500 m/z. The individual constituents obtained after GC were identified by comparing their mass spectra (MS) with standard library of National Institute for Standard Technology (NIST-07) mass spectra data.
Results and Discussion
In our study, selective enrichment method was employed to isolate chlorpyrifos degrading bacteria from apple orchard soil with previous history of chlorpyrifos usage. After 3 weeks of chlorpyrifos selection pressure, only three bacterial isolates were isolated on mineral salt agar medium with 50 mg/l chlorpyrifos. Many researchers had reported the isolation of chlorpyrifos degrading bacteria and fungi from soil.12,13,26,2 Few researchers had also reported isolation of chlorpyrifos degrading bacteria from sludge and waste water from pesticide manufacturing units.15, 27,19
Effective chlorpyrifos degrader bacterial isolate was selected from the three isolates by growing them in MSM supplemented with different concentration (50-800 mg/l) of chlorpyrifos as carbon and energy sole source. Isolate AST2.2 was able to tolerate 800 mg/l of chlorpyrifos and showed growth up to 400 mg/l of chlorpyrifos and further this isolate was selected for further studies (Table I). Previous reported results of researchers also observed that bacteria can tolerate and grow at high concentration of chlorpyrifos.21,27
Table 1: Chlorpyrifos resistance profile of selected bacterial isolates at different concentrations of chlorpyrifos in mineral salt agar medium
|
Bacterial strain |
Concentration of chlorpyrifos in MSM agar (mg/l) |
||||
|
50 |
100 |
200 |
400 |
800 |
|
|
AST1.1 |
+ |
+ |
+ |
- |
- |
|
AST2.1 |
+ |
+ |
+ |
- |
- |
|
AST2.2 |
+ |
+ |
+ |
+ |
± |
+ = Growth, ± = Mottled growth, - = Absence of growth
Strain AST2.2 colonies were creamish in color, circular shaped with smooth texture and entire margin. Strain AST2.2 was gram-negative, rod shaped, strictly aerobic, non-spore forming and motile. Strain AST2.2 gave positive results in biochemical tests for catalase, oxidase, citrate utilization, urease, and fermentation of glucose, but negative for lactose and sucrose fermentation, indole utilization, gelatin liquefaction, casein hydrolysis, methyl red and Voges-Proskauer. Based on various morphological and biochemical characteristics, the bacterial isolate ASK3.2 was identified as member of the genus Pseudomonas by standard protocol set in Bergey’s Manual of Determinative Bacteriology.28 This was further confirmed by partial sequencing of 16S rRNA gene (1231bp) and BLASTn analysis, the results of which showed more than 99 % similarity with species of Pseudomonas. 16S rRNA sequencing had been effectively used in molecular characterization of bacterial species. Researchers have used 16S rRNA sequencing for characterization of chlorpyrifos degrading bacterial isolates.12,10,29,27
EzTaxon was used for analysis of 16S rRNA gene sequences to achieve identification of isolates based on pairwise nucleotide similarity values and phylogenetic inference methods.30,22 Using EzTaxon server 16S rRNA sequence of strain ASK3.2 was identified as Pseudomonas resinovarans as its 16S rRNA sequence exhibited 97.70 % similarity with Pseudomonas resinovorans LMG 2274(T) (Accession no. Z76668). The sequencing result for AST2.2 was submitted to the GenBank National Center for Biotechnology Information (NCBI) database and the accession number obtained was KP322753.1. A dendrogram illustrates the results 16S rRNA analysis using PHYLIP of Fig. 1.
 |
Figure 1: Phylogenetic relationship of Pseudomonas resinovarans AST2.2 based on partial 16S rRNA sequence |
Pseudomonas resinovarans strain AST2.2 exhibited both extracellular and intracellular organophosphorus hydrolase activity. Extracellular OPH activity was 0.157 U/ml after 48 hrs while intracellular OPH activity was recorded 0.068 U/ml only. Our results fall in line with those of Ningfeng et al., 2004 [24] who reported OPHC2 activity 0.208 U/ml and 0.198 U/ml in Pseudomonas pseudoalcaligenes, isolated from OP-treated soil. This indicates OPH enzyme might be present in cell membrane or secreted extracellular by the strain AST2.2. Previous reported results indicate that OPHC2 enzyme was found to be located in cell membrane in Pseudomonas pseudoalcaligenes24 and OPH enzyme in Pseudomonas diminuta.31
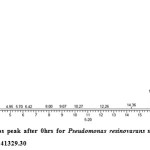
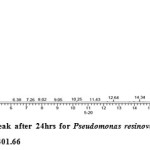
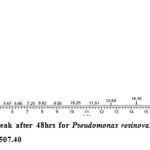
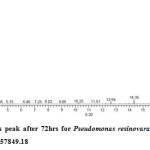
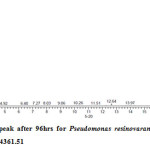
Gas chromatography equipped with flame ionization detector (FID) was employed for analysis of samples collected after interval of 24 hrs for 96 hrs using Shimadzu GC-17A gas chromatograph. GC studies were undertaken using liquid MSM and results are shown in Table II and figures II-VII, indicating percentage recovery of residual chlorpyrifos. Our results of chlorpyrifos degradation in mineral salt medium are in consonance with the results reported by Ajaz et al., 201218 where Pseudomonas putida MAS-1 was employed in degradation. It was found that with passage of time, the recovery of residual chlorpyrifos was gradually reduced.
Table 2: Percentage recovery of residual chlorpyrifos from mineral salt medium inoculated with Pseudomonas resinovorans strain AST2.2
|
Time (hrs) |
RT for Standard Chlorpyrifos |
Peak Height of standard chlorpyrifos |
RT for Sample |
Peak Height of sample |
Percent recovery of chlorpyrifos
|
Amount degraded (mg) |
Percent degraded |
|
0 |
16.03 |
542502.30 |
16.04 |
541329.30 |
99.78 |
0.018 |
0.22 |
|
24 |
16.03 |
542502.30 |
16.02 |
499401.66 |
92.00 |
0.640 |
8.00 |
|
48 |
16.03 |
542502.30 |
16.00 |
409507.40 |
75.40 |
1.968 |
24.60 |
|
72 |
16.03 |
542502.30 |
15.99 |
357849.18 |
65.96 |
2.723 |
34.03 |
|
96 |
16.03 |
542502.30 |
15.98 |
304361.51 |
56.10 |
3.512 |
43.90 |
In control sample (without any strain), standard chlorpyrifos peak was recorded at retention time RT 16.03 with peak height 542502.30. In test sample (inoculated with Pseudomonas strain AST2.2), chlorpyrifos peak was recorded at RT ranging from 16.04 to15.98 and peak height was found to decrease with increase in time, indicating decrease of chlorpyrifos amount in MSM. It was also observed that RT value decreases with increase in time (0-96 hrs). It was revealed that Pseudomonas resinovarans AST2.2 degraded chlorpyrifos rapidly and chlorpyrifos recovery was 99.78 % after 0 hrs which was gradually reduced to 56.10 % from MSM after 96 hrs. It was observed that the strain AST2.2 was not able to utilize/degrade chlorpyrifos during first 24 hrs when grown in MSM (Table II) as strain may be in earlier growth phases after inoculation into medium. Pseudomonas resinovarans AST2.2 degraded 43.90 % of 400 mg/l of chlorpyrifos after 96 hrs and this strain uses chlorpyrifos as substrate which also serve as source of carbon and energy.
Enterobacter strain B-14 degraded 40 % of 25 mg/l chlorpyrifos within 48 h,12 Stenotrophomonas sp. YC-1 degraded 100% of 100 mg/l within 24 hrs.10 Bacillus pumilus strain degraded 50 mg/l chlorpyrifos within 5 days of incubation, leaving behind 9 mg/l of pesticide17 and Synechocystis sp. strain PUPCCC 64 degraded 93.8 % of 5 mg/l chlorpyrifos within 5 days,32 these strains were able to hydrolyze chlorpyrifos to TCP.
 |
|
 |
|
 |
|
 |
|
 |
|
 |
|
Chlorpyrifos Intermediates Determination
After degradation studies, the samples were analyzed by using gas chromatographic-mass spectrometry (GC-MS) technique. The total ion current chromatograms were recorded between 40-500 m/z, with 965 scans. Both control and test sample resulted in similar fragment ions (96.9, 196.9, 285.9, 313.9 m/z). The GC-MS analysis exhibited the presence of chlorpyrifos at R.T. of 11.13 mins in control sample (Fig.VIII). Li et al., 2010 [25] determined 23 organophosphorus pesticides in surface water employing solid-phase micro extraction in combination with gas chromatography-mass spectrometry detection (GC-MS) and had reported chlorpyrifos peak at retention time (RT) 11.41±0.01. On comparison with standard library of NIST-07 mass spectral database confirmed the matching of mass/charge ratio versus relative intensity at R.T 11.11 for the test sample to standard spectra of chlorpyrifos (Fig.IX). One probable metabolite i.e. phosphoric acid, diethyl 3,5,6-trichloro-2-pyridyl ester (C9H11Cl3NO4P) with CAS no. 5598-15-2 was identified by NIST library search along with chlorpyrifos in test sample at RT 11.11. The mass spectra results obtained from GC-MS analysis exhibited that chlorpyrifos was degraded further into smaller intermediates which were not be identified employing the available library database. The presence of chlorpyrifos was observed at R.T. 11.11 min but no other intermediate was determined till R.T. 15 min. Results obtained infer that chlorpyrifos was possibly completely metabolized by the isolates into smaller intermediates. Kumar 2011,33 and Barathidasan et al., 2014,34 also reported similar results after GC-MS analysis as smaller intermediates after chlorpyrifos degradation were not identified.
 |
|
 |
|
Conclusion
For the purpose of ecological restoration, bioremediation/biodegradation based techniques are gaining popularity. Pseudomonas resinovarans strain AST2.2 can be used for bioremediation of contaminated soils with chlorpyrifos
Acknowledgements
The author acknowledges the financial support from the Dean, College of Horticulture, Dr Y.S. Parmar University of Horticulture and Forestry, Solan (H.P.).
References
- Gallo M A and Lawryk N J 1990. The Handbook of Pesticide Toxicology. CA: Academic Press, San Diego, 920 – 925.
- Silambarasan S and Abraham J 2013. Ecofriendly Method for Bioremediation of Chlorpyrifos from agricultural soil by novel Fungus Aspergillus terreus Water Air Soil Pollution 224: 1364 - 1369.
- Swati and Singh D K 2002. Utilization of chlorpyrifos by Aspergillus niger and flavus as carbon and phosphorus source. 17th World Congress of Soil Science, 14 - 21 Aug 2001, Bangkok, Thailand
- Qiao C L, Yan Y C, Shang H Y, Zhou X T and Zhang Y 2003. Biodegradation of pesticides by immobilized recombinant Escherichia coli. Bulletin of Environmental Contamination and Toxicology 71: 370 - 374.
- Environmental Protection Agency. Review of chlorpyrifos poisoning data. Environmental Protection Agency, Washington, D.C.
- Racke KD, Coats JR, Titus KR (1988) Degradation of chlorpyrifos and its hydrolysis products, 3,5,6-trichloro-2- pyridinol, in soil. Journal of Environmental Science and Health B23: 527 – 539.
- Whitacre D M 2012. Reviews of Environmental Contamination and Toxicology. Springer Dordrecht Heidelberg, London. New York.
- Munnecke D M and Hsieh D P H 1974. Microbial decontamination of parathion and p-nitrophenol in aqueous media. Applied and Environmental Microbiology 28: 212 – 217.
- Mallick B K, Banerji A, Shakil N A amd Sethunathan N N Bacterial degradation of chlorpyrifos in pure culture and in soil. Bulletin of Environmental Contamination and Toxicology 62: 48 – 55.
CrossRef - Yang C, Liu N, Guo X and Qiao C 2006. Cloning of mpd gene from a chlorpyrifos-degrading bacterium and use of this strain in bioremediation of contaminated soil. FEMS Microbiology Letters 265: 118 – 125.
CrossRef - Ohshiro K, Kakuta T, Sakai T, Hirota H, Hoshino T and Uchiyama T 1996. Biodegradation of organophosphorus insecticides by bacteria isolated from turf green soil. Journal of Fermentation Bioengineering 82: 299 - 305.
- Singh B K, Walker A, Morgan J A W and Wright D J 2004. Biodegradation of chlorpyrifos by Enterobacter strain B-14 and its use in bioremediation of contaminated soils. Applied and Environmental Microbiology 70(8): 4855 – 4863.
- Yang L, Zhao Y H, Zhang B X, Yang C H and Zhang X 2005. Isolation and characterization of a chlorpyrifos and 3,5,6-trichloro-2-pyridinol degrading bacterium. FEMS Microbiology Letters 251(1): 67 – 73.
- Ghanem I, Orfi M and Shamma M 2007. Biodegradation of chlorpyrifos by Klebsiella isolated from an activated sludge sample of waste water treatment plant in Damascus. Folia Microbiologica 52(4): 423 - 427.
- Li X H, He J and Li S P 2007. Isolation of a chlorpyrifos-degrading bacterium, Sphingomonas strain Dsp-2, and cloning of the mpd gene. Ressearch Microbiology 158(2): 143 – 149.
CrossRef - Fulekar M H and Geetha M 2008. Bioremediation of Chlorpyrifos by Pseudomonas aeruginosa using scale up technique. Journal of Applied Biosciences 12: 657 – 660.
- Anwar S, Liaquat F, Khan Q M, Khalid Z M and Iqbal S 2009. Biodegradation of chlorpyrifos and its hydrolysis product 3,5,6-trichloro-2-pyridinol by Bacillus pumilus strain C2A1. Journal of Hazardous Materials 168(1): 400 – 405.
- Ajaz M, Rasool S A, Sherwani S K and Ali T A 2012. High Profile Chlorpyrifos Degrading Pseudomonas putida MAS-1 from Indigenous Soil: Gas Chromatographic Analysis and Molecular Characterization. International Journal of Basic Medical Sciences and Pharmacy 2(2): 58 - 61.
- Lu P, Li Q, Liu H, Feng Z, Yan X, Hong Q and Li S 2013. Biodegradation of chlorpyrifos and 3,5,6-trichloro-2-pyridinol by Cupriavidus DT-1. Bioresource Technology 127: 337 - 342.
- Singh S and Singh D K (2003) Utilization of monocrotophos as phosphorus source by Pseudomonas aeruginosa F10B and Clavibacter michiganense insidiosum SBL 11. Canadian Journal of Microbiology 49: 101 – 109.
- Bhagobaty R K and Malik A 2008. Utilization of Chlorpyrifos as a sole source of carbon by bacteria isolated from wastewater irrigated agricultural soils in an industrial area of western Uttar Pradesh, India. Research Journal of Microbiology 3(5): 293 - 307.
- Kim O S, Cho Y J, Lee K, Yoon S H, Kim M, Na H, Park S C, Jeon Y S, Lee J K, Yi H, Won S and Chun J 2012. Introducing EzTaxon-e: a prokaryotic 16S rRNA gene sequence database with phylotypes that represent uncultured species. International Journal of Systematic & Evolutionary Microbiology 62: 716 - 721.
- Thompson J D, Gibson T J, Plewniak F, Jeanmougin F and Higgins D J 1997. The clustral X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acid Research 24: 4876 - 4882.
CrossRef - Ningfeng W U, Minjie D, Xiuyun S, Guoyi L, Bin Y and Yunliu F 2004. Isolation, purification and characterization of a new organphosphorus hydrolase OPHC2. Chinese Science Bulletin 49(3): 268 - 272.
- Li X, Gan P, Peng R, Huang C and Yu H 2010. Determination of 23 Organophosphorous Pesticides in Surface Water Using SPME Followed by GC–MS. Journal of Chromatographic Science 48: 183 - 187.
CrossRef - Ajaz M, Jabeen N, Akhtar S and Rasool S A 2005. Chlorpyrifos resistant bacteria from Pakistani soil: isolation, identification, resistance profile and growth kinetics. Pakistan Journal of Botany 37(2): 381 - 388.
- Latifi A M, Khodi S, Mirzaei M, Meresmalli M and Babavalian H 2012. Isolation and characterization of five chlorpyrifos degrading bacteria. African Journal of Biotechnology 11(13): 3140 - 3146.
- Holt J G, Krieg N R, Sneath P H, Staley J T and Williams S T 1994. Bergey’s Manual of Determinative Bacteriology.
- Rani M, Surekha L K, Vijaya D P, Suvarnalatha M J R, Aruna J S K, Narasimha G and Venkateswarlu K 2008. Isolation and characterization of a chlorpyrifos degrading bacterium from agricultural soil and its growth response. African Journal of Microbiology Research 2: 026 - 031.
- Chun J, Lee J K, Jung Y, Kim M, Kim S, Kim B K and Lim Y M 2007. EzTaxon: a web-based tool for the identification of prokaryotes based on 16S ribosomal RNA gene sequences. International Journal of Systematic and Evolutionary Microbiology 57: 2259 – 2261.
CrossRef - Dumas D, Caldwell S R, Wild J R and Raushel F M 1989. Purification and Properties of the Phosphotriesterase from Pseudomonas diminuta. The Journal of Biological Chemistry 264(33): 19659 - 19665.
- Singh D P, Khattar J I S, Nadda J, Singh Y, Garg A, Kaur N and Gulati A 2011. Chlorpyrifos degradation by the cyanobacterium Synechocystis strain PUPCCC 64. Environmental Science and Pollution Research 18(2): 1351 – 1359.
CrossRef - Kumar S 2011. Bioremediation of chlorpyrifos by bacteria isolated from the cultivated soils. International Journal of Pharma and Bio Sciences 2(3): 359 - 366.
- Barathidasan K, Reetha D, Milton D J, Sriram N and Govindammal M 2014. Biodegradation of chlorpyrifos by co-culture of Cellulomonas fimi and Phanerochaete chrysosporium. African Journal of Microbiology 8(9): 961 - 966.
CrossRef







