Metaanalysis of Public Wastewater Metagenomes: Revealing the Influence of Climatic Variations on the Abundance of the Bacterial Members
1
Department of Microbiology,
St. Xavier’s College (Autonomous), Kolkata,
Mother Teresa Sarani, Mullick Bazar, Park Street area,
Kolkata,
West Bengal
India
Corresponding author Email: mmgxaviers@gmail.com
DOI: http://dx.doi.org/10.12944/CWE.19.1.11
Copy the following to cite this article:
Karmakar R, Mondal K, Ghosh M. M. Metaanalysis of Public Wastewater Metagenomes: Revealing the Influence of Climatic Variations on the Abundance of the Bacterial Members. Curr World Environ 2024;19(1). DOI:http://dx.doi.org/10.12944/CWE.19.1.11
Copy the following to cite this URL:
Karmakar R, Mondal K, Ghosh M. M. Metaanalysis of Public Wastewater Metagenomes: Revealing the Influence of Climatic Variations on the Abundance of the Bacterial Members. Curr World Environ 2024;19(1).
Download article (pdf) Citation Manager Publish History
Select type of program for download
| Endnote EndNote format (Mac & Win) | |
| Reference Manager Ris format (Win only) | |
| Procite Ris format (Win only) | |
| Medlars Format | |
| RefWorks Format RefWorks format (Mac & Win) | |
| BibTex Format BibTex format (Mac & Win) |
Article Publishing History
| Received: | 2024-02-09 |
|---|---|
| Accepted: | 2024-04-25 |
| Reviewed by: | 
 Grigorios Kyriakopoulos
Grigorios Kyriakopoulos
|
| Second Review by: |

 Aleksandar Racz
Aleksandar Racz
|
| Final Approval by: | Dr. Wilkister Nyaora Moturi |
Introduction
A long-term alteration in the Earth's climate system, manifesting as novel weather patterns, has been linked to the concurrent development of contemporary civilization, particularly the processes of urbanization and industrialization.1 For example, anthropogenic greenhouse gas emissions have triggered post-industrial climate warming, resulting in each of the preceding few decades being the warmest on record. If the current trends continue, by 2040, Earth may confront a global temperature surge of about 1.5 ° C.2 With the rising temperature, environmental; ecological and economic consequences of climatic shifts have been have found to be reflected in multiple domains. Glacier shrinkage, global increase in Earth's average surface temperature, greater precipitation, higher concentrations of greenhouse gases in ocean systems are few of them.3
Moreover, incidence of various infectious diseases together with the evolution and spread of multi-drug resistant pathogens through wastewater network, appear as the two most vulnerable aspects of the ongoing climate crisis. The advent of Antimicrobial Resistance (AMR) in environmental microbial members is not novel. Microbial communities exhibit a well-documented potential to synthesize a diverse array of metabolites structurally analogous to the established antibiotics employed in human healthcare. However, the recent surge in synthetic antibiotics in medicine has fundamentally changed the landscape for how resistance mechanisms evolve in microbes. This transformation is mainly attributed to the substantial selection pressures imposed, particularly on the microbial communities of humans and domestic livestock, as well as in the ecological niches that are heavily contaminated with antibiotics. The aforementioned selective pressures have demonstrably facilitated the promoted the transfer and spread of a diverse array of antibiotic resistance genes (ARGs) across varied bacterial taxa including the pathogenic ones.4
Exacerbating the issue of antibiotic overuse, inadequately managed wastewater infrastructure followed by the overflow of community sewage and agricultural effluent contribute to the proliferation of multi-drug resistant microbial members. These are the two promising routes through which flooding facilitates dissemination of multidrug resistant pathogenic microbes into the environment. An investigation by Liu et al. (2017) identified a statistically significant correlation between flood intensity and shigellosis incidence in Baise, China. Their study, encompassing the period 2004-2012, demonstrated that both moderate and severe flood events acted to exacerbate the comparative risk of shigellosis. In addition to this previous disease disaster there is a report which is much more concerning- as per IPCC report. 5,6 global temperature hike of 20 C can increase the frequency of natural calamities like drought, flooding, extreme precipitation, storms etc, reducing the accessibility of safe food and drinking water. Beyond floods, water scarcity induced by drought presents a significant risk factor. In due course of limited water availability, communities are forced to share resources, inevitably compromising sanitation practices which in turn creates an environment conducive to the spread of various waterborne illnesses. The upsurge of dysentery caused by the broad spectrum multi-resistant E. coli in Africa serves as a stark illustration of the detrimental consequences associated with inadequate sanitation practices.7 On the other hand, research suggests a potential link between rising ambient temperatures and increased antibiotic resistance in prevalent nosocomial infections. A study documented a corresponding elevation in resistance among specific pathogens viz. Escherichia coli, Klebsiella pneumoniae, and Staphylococcus aureus by 4.2%, 2.2%, and 2.7% respectively, following a ten-degree Celsius temperature increase across the United States.8 Moreover, according to a recent study, spread of several Gram-negative bacterial infections is directly linked to humidity, temperature and monthly precipitation.9 Fluctuations in environmental conditions can even act as a catalyst for the emergence of novel transposable genetic determinants attributed to resistance mechanism. Notably, research has shown a significant influence of local ambient temperature on the lateral transfer of resistance genes like the New Delhi metallo-?-lactamase 1 (NDM-1) and OXA-48-like carbapenemases.10 Similarly, Sweileh, 2020 have studied that the prevalence of few gastrointestinal as well as respiratory infections caused by Shigella sp., and Campylobacter sp. varies with variation in seasonal temperature and precipitation.1
Influence of climate change in wastewater network
The evolving climatic regime presents significant challenges to the operational efficiency and resilience of wastewater networks which is quite evident and may be noted in following few domains-
Changing climatic circumstances alter the quantity and quality of water that is available throughout time and place. This has an impact on how people use water, which in turn affects sanitation and contributes to the spread of infectious illnesses.11
Combined Sewage Overflow that is the uncontrolled sewage discharge and spills induced by heavy rainfall facilitates increased influx of microbial pathogens including the nosocomial ones into the receiving environment presenting a higher risk of infection such as faecal-oral disorders, vector-borne illnesses, toxicity, cirrhosis, dermatitis, etc. Rapid entrance of antimicrobial resistance determinants into the food chain through wastewater contaminated community drinking water is also a concerning matter.11
Being the repository for majority of the pathogenic microbial members, vulnerability of wastewater systems to the climatic variation can put public health at stake. Antibiotic resistant bacteria (ARB) are widely spreading in the ecosystem through contaminated wastewater, which is a prelude to the resistome cycle. According to estimates, polluted drinking water is responsible for about 4,85,000 global annual deaths from diarrhoea.12
Changes in climate also requires adaptation, in terms of how wastewater is managed. Better understanding of influences of climate change is a mandate in this domain for wastewater sector decision-makers to feel more confident when creating policies and engaging in adaptive planning.
The last domain needs a periodic monitoring of wastewater samples accumulating community sludges which in turn can put a light on the distribution and abundance of microbial members in the wastewater environment. This in turn can aid in foretelling the disease threat and unveiling the trend of antibiotic resistance in the community.
Community Genomics in large-scale monitoring wastewater monitoring:
Metagenomic mapping of the wastewater samples followed by downstream meta-analysis have been proved to effectively aid this large-scale monitoring of the community sewer network. Taking the effectiveness of wastewater-based epidemiology into consideration, this study focusses in complete profiling of wastewater bacteria and prediction of pathogenic load through culture-independent Metagenomic Mapping of wastewater samples collected from various sewer networks in Newcastle (United Kingdom); Zimbabwe (Africa) and West Bengal (India). Detection of spatial variation as well as degree of commonness in wastewater bacterial profile across the three samples collected from the three above mentioned countries is another area that has been studied. This approach is mandatory to assess pathogenic load, predicting upcoming disease outbreaks and to raise community awareness of sanitation measures in response.
Materials and Methods
Data Description
Three distinct wastewater metagenomic data were collected form Sequence Read Archive (SRA) available at NCBI database. Accession numbers of the samples under study includes- SRA: ERS946133, SRA: ERS613823 and SRA: SRS4735877. Information regarding each of the three metagenomics datasets has been presented in a tabular format in Table 1.
Table 1: Chosen metagenomic datasets corresponding to different wastewater samples collected from NCBI SRA
Sample Nomenclature | SRA Details | Date of submission | Region of isolation | Wastewater Type |
Sample 1 | ERS946133 | 27/10/2018 | Newcastle University, United Kingdom. | Wastewater from sewer network |
Sample 2 | ERS613823 | 29/04/2015 | Zimbabwe, Africa | Beer plant wastewater sample |
Sample 3 | SRS4735877 | 08/10/2018 | West Bengal, India | Sludge from urban sewage |
Rationale for data point selection
Metagenomic datasets generated through Illumina platform-based amplicon sequencing of three distinct wastewater samples having three distinct regions of isolation were retrieved from NCBI database.
Sample 1 represented the wastewater collected from a sewer network of Newcastle University, United Kingdom. The wastewater management status in the United Kingdom is generally considered to be robust and well-regulated. The country has invested significantly in wastewater treatment infrastructure over the years, resulting in high standards of water quality and environmental protection. While substantial hurdles including deteriorating infrastructure, burgeoning demographics, and the imprudent use of antibiotics in livestock production resulting in a high incidence of drug-resistant microbial infections affecting an estimated 60% of the population, considerably challenges the wastewater management cycle of the country. Furthermore, the United Kingdom leads in investigating emerging pollutants found in wastewater, such as pharmaceuticals, personal care products, and microplastics.13 Therefore, evaluating wastewater health within this specific framework can offer valuable insights into the potential environmental and public health implications, even in well-managed wastewater systems.
Sample 2 was the Beer plant wastewater sample collected from Zimbabwe, Africa. Approximately 63% of this landlocked country situated in Southern Africa grapples with insufficient rainfall, resulting in water scarcity. Consequently, the government is compelled to resort to wastewater reuse and recycling for purposes such as irrigation, industrial use, and other non-potable needs. Furthermore, economic constraints hinder investment in conventional wastewater treatment infrastructure within the country.14 Therefore, gaining insights into the state of wastewater health in the country can serve as a valuable tool safeguarding public health and monitoring environmental well-being.
Sample 3 represented sewage sludge, collected from urban sewage of West Bengal, India. As the fourth most populous state in the nation, this state showcases significant diversity in demographics and climate, leading to a substantial burden of infectious diseases. The availability of over-the-counter antimicrobials, misuse of newer antibiotic classes, and inadequate wastewater management infrastructure further compound the challenges within the water management system of the state.15. Given the need to understand how microbial load and antibiotic resistance patterns vary across geographically diverse locations, West Bengal have emerged as a crucial site for data collection.
Metaanalysis of Metagenomic Data
Followed by quality checking, computational analysis of the raw metagenomic data was performed using various web servers and web tools.
Taxon calling and OTU clustering of the quality checked sequence reads, obtained from the amplicon sequencing was used for thorough profiling of microbial population using MetaG server, that imparts taxonomic attributes to individual reads.16
In next step, a suitable interactive tool- Venny 2.1.0 was used to identify a collection of common and unique genera throughout the test samples.17
Following identification, metagenome-wide association studies were conducted to link microbes in the sample to the host disorders in order to predict pathogenic load in the community and to understand disease networks of microbial members using Taxon Set Enrichment Analysis (TSEA).18
In the next step, 16S microbiome data was then leveraged to predict the core enriched metabolic functions within the microbial community. Additionally, function-driven correlation networks were constructed to elucidate the interaction patterns between these microbes. Global Mapper Module was utilised to examine the functional attributes of the microbial consortium, focusing on the presence of various metabolic pathways.19
The overall methodology of the particular study had been summarised and represented in form of a Conceptual Framework labelled as Figure 1.
 | Figure 1: Conceptual Framework of the study
|
Results
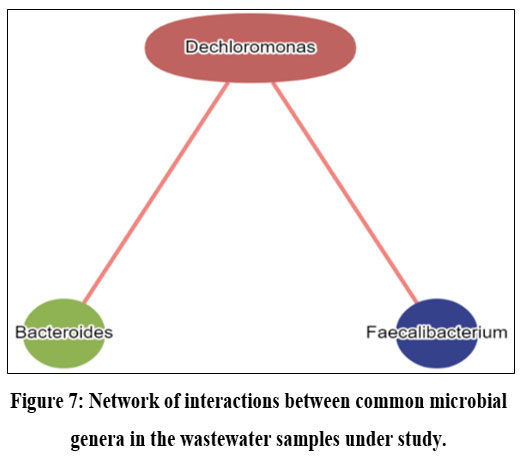
Computational analysis of metagenomic data revealed that alpha diversity in the wastewater samples collected from United Kingdom (Sample 1); Africa (Sample 2) and India (Sample 3) was 39, 52 and 124 respectively. Amidst them, Desulfitobacterium, Prevotella and Sulfurospirillum were prevalent (23%, 34% and 40% respectively) in sample 1, 2 and 3 respectively. Whereas Enhydrobacter, Renibacterium and Burkholderia were respectively found to have lowest abundances in the 3 samples. Few members of the human gut microbiome, including Prevotella and Bacteroides, were ubiquitously found in wastewater samples from Africa and India along with the additional prevalence of Pseudomonas and Burkholderia, known for their association with hospital-acquired infections. Interestingly, bacterial genera like Desulfitobacterium, Sulfurospirillum, and Desulfovibrio, posing a substantial role in the sulfur cycle and anaerobic metabolism across various environments, were consistently present in all the samples analysed (Figure 2A; 2B; 2C). Comparison of wastewater bacterial profile revealed that sample 1, sample 2 and sample 3 respectively contained 10.4%, 11% and 56.1% of unique bacterial genera while only 3 bacterial genera- Bacteroides, Dechloromonas and Faecalibacterium were common throughout them (Figure 2D).
Wastewater analysis in Africa and India exhibited a greater abundance of Bacteroides compared to United Kingdome. This trend could potentially be attributed to the moderate temperature range (35-37°C) prevalent in these South Asian nations, which aligns with optimal growth conditions for this bacterial genus. Whereas, Dechloromonas showed predominance in the sludge from India followed by in United Kingdom. The abundance of Faecalibacterium exhibited minimal variation across the three samples, with a modest enrichment observed in the African sample. This observed pattern may be attributable to the mesophilic temperature range shared by all three countries, which coincides with the optimal growth conditions documented for this bacterial genus (Figure 3).
Detection of pathogenic load within common bacterial genera through taxon set analysis suggests an increased likelihood of multiple illnesses occurring within the network depicted in Figure 3. Liver cirrhosis, irritable bowel syndrome, ulcerative colitis, Diarrhoea were some of the noticeable disorders associated with ill managed wastewater. Health complications like Type 1 Diabetes, Colorectal cancer was among the secondary infections that may arise (Figure 4)
Figure 5A and 5B depicts pathogenic load of unique bacterial genera present in sample 1 and sample 2 respectively. Flavobacterium in sample 1 had been predicted to cause Ulcerative Colitis whereas Clostridium in sample 2 was found to be associated to Hepatitis B only. In contrast to these, bacterial genera in sample 3 showed high pathogenic load and disease association potency, as evidenced by the diseases like Liver Cirrhosis, Colitis, Hepatitis B, Bacterial Vaginosis, periodontitis in the disease network shown in figure 5C. Noticeable pathogenic bacterial genera associated to these disorders were found to be Prevotella, Clostridium, Pseudomonas, Escherichia, Klebsiella, Streptococcus and Acinetobacter.
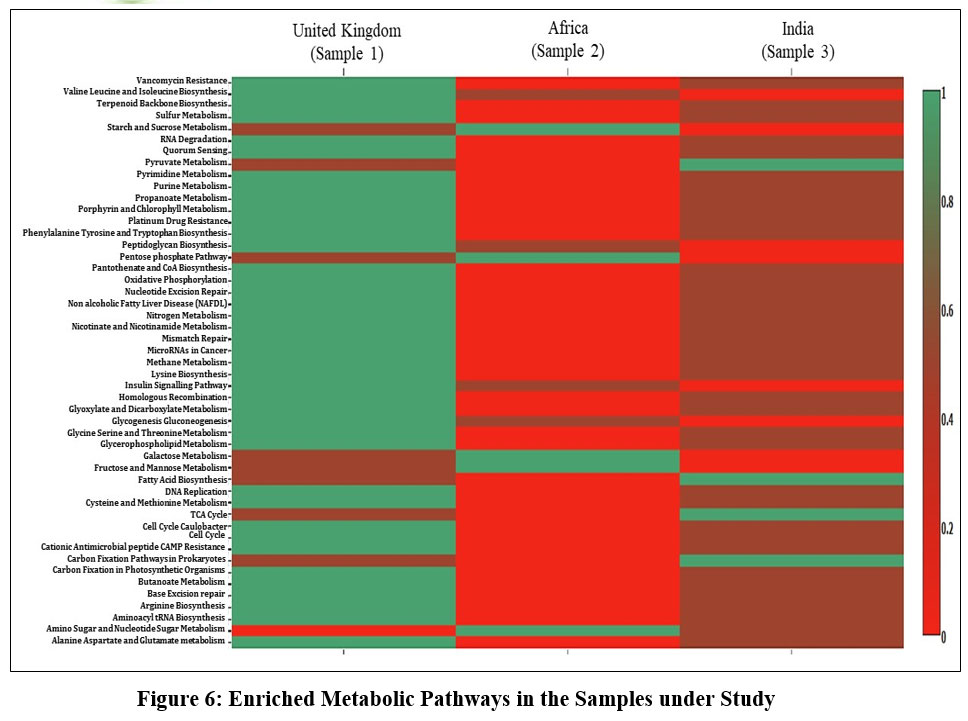
Core pathway analysis of the microbial community revealed predominance of few important resistance pathways like - Vancomycin resistance pathway, Platinum drug resistance pathway in sample 1. Whereas sample 2 expressed none of these resistance pathways. Similar trend was observed in case of nitrogen, methane and sulfur metabolism pathways as well as even in case of lysin, tyrosine, tryptophan biosynthesis pathway. In contrast, multiple sugar and nucleotide metabolic pathways showed higher expression in sample 2 and intermediate to no expression in sample 1. Wastewater metagenome from sample 3 was found to show intermediate expression of vancomycin, drug resistance pathway as well as various sugar metabolism pathways. But interestingly prokaryotic specific carbon fixation pathway was predominant only in sample 3 followed by little and no expression in sample 1 and sample 2 respectively. (Figure 6). Wastewater microbial network analysis of common bacterial genera across the three samples revealed Dechloromonas has unidirectional relationship with Bacteroides and Faecalibacterium which is represented through the red line (Figure 7).
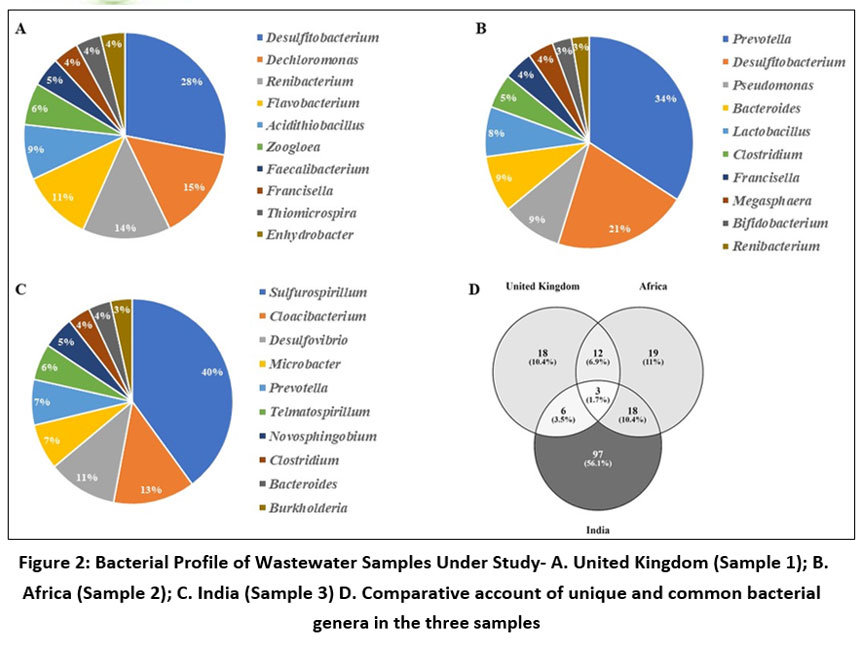 | Figure 2: Bacterial Profile of Wastewater Samples Under Study- A. United Kingdom (Sample 1); B. Africa (Sample 2); C. India (Sample 3) D. Comparative account of unique and common bacterial genera in the three samples
|
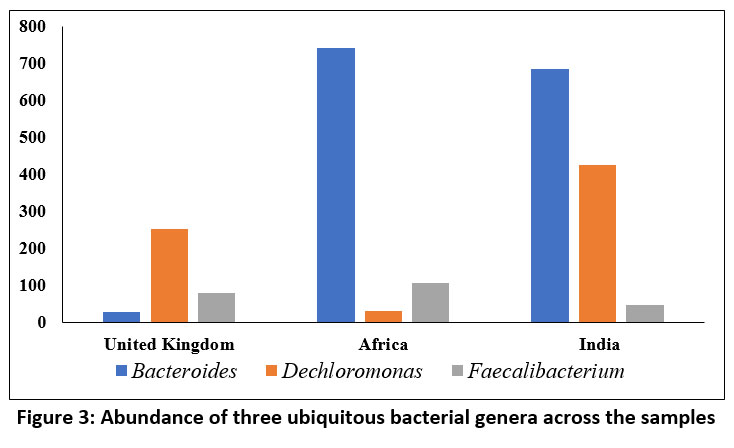 | Figure 3: Abundance of three ubiquitous bacterial genera across the samples
|
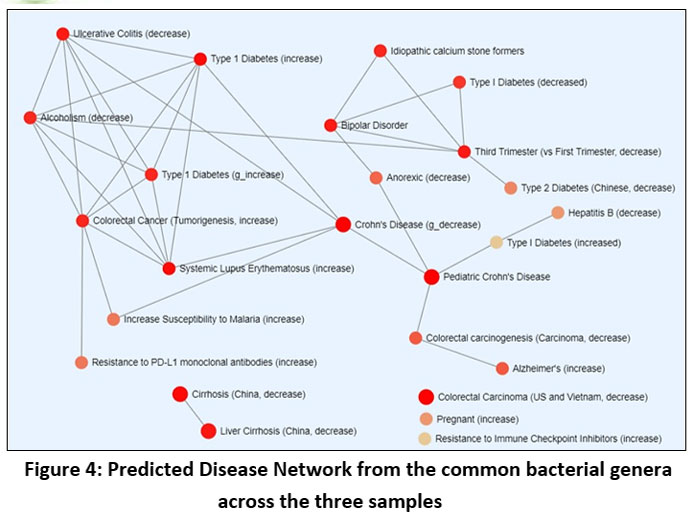 | Figure 4: Predicted Disease Network from the common bacterial genera across the three samples
|
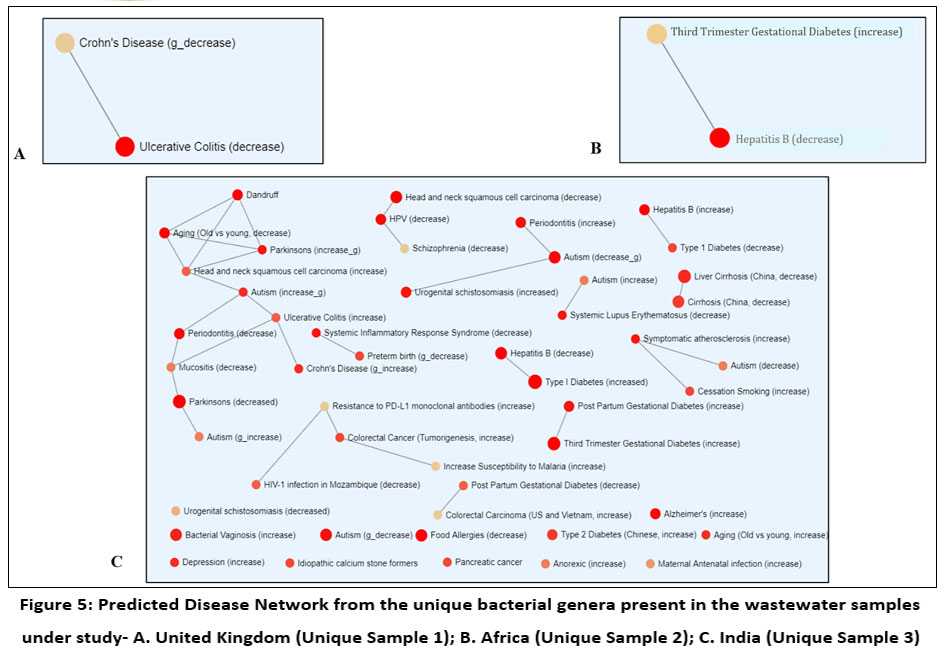 | Figure 5: Predicted Disease Network from the unique bacterial genera present in the wastewater samples under study- A. United Kingdom (Unique Sample 1); B. Africa (Unique Sample 2); C. India (Unique Sample 3)
|
 | Figure 6: Enriched Metabolic Pathways in the Samples under Study
|
 | Figure 7: Network of interactions between common microbial genera in the wastewater samples under study.
|
Discussion
Climate change has cemented its status as an unavoidable occurrence in the recent years, and it is predicted to continue to intensify the frequency, severity, and effects of several types of extreme weather occurrences in the years ahead. On the other hand, anthropogenic release of multiple greenhouse gases has caused global average temperature to elevate since the middle of the 19th century. The observed phenomenon of gradual climate warming has been demonstrably associated with the re-emergence and intensified geographic spread of a diverse range of infectious diseases. This association can be attributed to the dependence of these vector-borne, waterborne, airborne, and foodborne pathogens on environmental factors for their viability and transmission cycles. The escalating incidence of contagious diseases compels a reliance on increased antimicrobial treatment regimens. However, this very practice inadvertently fuels the emergence of antibiotic resistance. The widespread use of antibiotics followed by their discharge, often untreated, into the environment fosters the creation of reservoirs harbouring antibiotic-resistant bacteria. Moreover, increase in sea level as well as in surface temperature have also been found to induce growth of Vibrio cholerae, Vibrio vulnificus in Baltic Sea and North Sea during the hot summer of 2006. Thus, the escalating phenomenon of climate change can be demonstrably linked to the emergence of AMR which itself constitutes a critical and existential threat to global public health, necessitating the implementation of coordinated international efforts to mitigate this growing challenge.20 Temperature and pH, among other environmental parameters, significantly influence the diverse mechanisms employed by pathogenic microbes for adaptation to a changing ecological landscape. A 2020 study by Kaba et al. conducted in Göttingen, Germany, has highlighted a seasonal variation in the prevalence of antibiotic-resistant bacteria, with a higher incidence of Carbapenem-resistant Pseudomonas aeruginosa, and Methicillin-resistant Staphylococcus aureus observed in warmer months. 21 Ecological parameters pose a decisive impression in the emergence of AMR amongst pathogenic bacteria. The discharge of inadequately treated industrial effluents, pharmaceutical waste, and hospital wastewater into water sources serves as a significant driver of AMR. These effluents introduce not only resistant bacterial strains but also selective pressures that promote the proliferation of resistant phenotypes within aquatic ecosystems. A particularly concerning example is a study examining AMR in the Cauvery River, Karnataka, which identified 100% of isolated Escherichia coli exhibiting resistance to third-generation cephalosporins. This finding underscores the alarming prevalence of AMR within this critical water resource.22 In this context, wastewater management simply cannot be overlooked as it plays critical societal functions that is impacted by the current climate change situation. One immediate impact includes extreme weather events induced malfunctioning of sewage treatment plant which in combination with sewage overflow cause contamination of freshwater aquifers leading to serious health hazards. Thus, in order to shed light on the impending disease threats in a community, based on the dispersion and abundance of microbial members in its sewer network, routine monitoring of wastewater samples accumulating community sludge is a mandate.
Taking into account on the said context, detection of spatial variation as well as degree of commonness in wastewater bacterial profile and disease burden across the three wastewater samples collected from three different locations namely- Newcastle (United Kingdom); Zimbabwe (Africa) and West Bengal (India) have been made possible in this study.
Upon examining the pattern of bacterial overabundance in each of the wastewater samples under study, the genus, Desulfitobacterium was found to be most prevalent across our sample 1, that is from United Kingdom. Here, in the United Kingdom, the average temperature ranges from 25°C to 32.8°C presenting optimum growth condition for the aforesaid bacterium.23 Whereas, Prevotella exhibited highest abundance in the sample from Africa (Sample 2). One interesting fact about Prevotella is that they have been found predominantly in those individuals who intake more carbohydrate, particularly fibre, instead of protein and animal fats.24 It has been reported that in Africa, crops like grains including cereals, corn, and barley, and tubers like sweet potatoes that are less impacted by inclement weather gradually gained popularity throughout the continent making the continental diet largely dependent on the grains.25 All of these facts strongly suggest the reason behind the prevalence of the particular genera in African region. Similarly, predominance of Sulfurospirillum in wastewater sample of India (Sample 3) can be corelated with warm and humid climate of the country. A study by Kumar et al., 2021 have highlighted the prevalence of Sulfurospirillum in the textile industrial effluent in an around Gujarat, India.26
In context to the unique bacterial members growing in the each and individual study sites, it is noteworthy that sample 1 has a unique occurrence of Flavobacterium which is related to the mesophilic nature of that specific genus that makes it able to flourish in the climatic condition of Newcastle, United Kingdom.27 On the other hand, the occurrence of Clostridium members is linked to their spores' capacity to withstand the harsh climate of Zimbabwe, Africa, which elucidates the relationship between Clostridium prevalence.28 Escherichia, the uniquely obtained bacterial genus from the sample 3 is linked to the optimum temperature range of Indian climate favouring the growth of these two mesophilic bacterial members.29
A comprehensive analysis of the level of similarity in bacterial load among the samples being studied, three bacterial genera viz. Bacteroides, Dechloromonas, and Faecalibacterium emerged as notable. All these genera being mesophiles got better survival advantage in both the tropical and temperate climate. Feaclibacterium and Bacteroides, that were found across the samples, have been associated with diarrhoeal disease which occurs frequently in monsoon season.
Regarding the prediction of pathogenic load associated with the bacterial consortia, it is noteworthy to observe that Faecalibacterium, a member present ubiquitously in all three samples under investigation, exhibits an inverse association with the incidence of diarrheal disease concerning its abundance within an environmental niche. Conversely, a positive correlation is observed between diarrhoeal disease and the abundance of Bacteroides.30 It must be mentioned that the only strain of Bacteroides spp. linked to diarrheal illness is Enterotoxigenic Bacteroides fragilis (ETBF), or B. fragilis toxin-producing strains.31 Typically, the Indian summer monsoon season runs from June to September and the degree of precipitation during this period is 2000 mm (78 inches) whereas in Zimbabwe (Africa), precipitation occurs in in two distinct periods- one from October to December and other from January to March.32,33 These typical climatic features facilitate poor sanitation and contamination of freshwater resources through community sewage overflow which are in turn linked to frequent diarrhoeal incidence in 3rd world countries like India and Africa.34 Results of enrichment analysis based pathogenic load prediction, also shed light on the fact that ulcerative colitis, another disease association identified and predicted, with the causative organism Bacteroides was found across the samples, occurs in greater number in UK in comparison with the two tropical regions, India and Africa.35 This type of result is supported by the report of Aamodt et al., 2013, which states that with the rise in summer time temperature by only one degree, occurrences of ulcerative colitis lower by 9%.36 Moreover, most of the diseases which were observed in the pathogenic load of common genera across the samples are significantly linked with Bacteroides and Feaclibacterium rather than Dechloromonas.
In a nutshell, through this particular study, it could be enlightened that how change in the temperature and rainfall is related to the differences in the microbial abundance across these three regions under study. This warrants further exploration to establish a definitive link between these environmental factors and microbial populations. Along with the change in the abiotic factors, it has also been evidenced from our study that climate-based change in the dietary pattern also drives great impact on the microbial diversity. On the other hand, in context of the disease occurrences, favourable temperature acts as potent driver behind various diseases. Also, the adaptability of some of the disease-causing microbes are commendable, as the climate is changing, for sake of survival, they are continuously adapting to the new climatic conditions.
Moreover, misuse of various broad-spectrum antibiotics and their rapid discharge into the water bodies from domestic, industrial, veterinary and agricultural sewer networks is an emerging health care concerns of 21st century. This practice accelerates the natural selection of genetic determinants within wastewater bacterial communities favouring the emergence of antibiotic resistance among these microbes.37 Through analysis of enriched metabolic pathways across the three samples, our study has also revealed the presence of vancomycin and platinum drug resistance pathway in sample 1 (United Kingdom), which in turn supports the above-mentioned facts. Our data supports the recent data by United Kingdom Health Security Agency which says three out of every five individuals over there suffers from drug resistant microbial infections which can be corelated with the over prescription as well as overuse of antibiotics in animal husbandry.38 Understanding the intricate interaction between climate change and aquatic diseases can thus guide measures to improve public health and environmental management.
Conclusion
If we look at the current scenario, waterborne diseases are becoming one of the main factors behind the occurrence of high morbidity, mortality, and substantial economic burdens worldwide. Efficient and prompt detection, along with effective monitoring of the spread of waterborne diseases, is crucial for early intervention and preventing the rapid dissemination of the same. Furthermore, clinical surveillance protocols present numerous constraints, including the invasive nature of testing procedures and challenges associated with testing a large population. In this context, sewage surveillance followed by metagenomic mapping of wastewater samples is proving its efficacies not only in finding spatial variations in microbial distribution across the wastewater systems globally but also in prediction of upcoming disease threat and extent of antibiotic resistance in a community. This particular study through its finding has proved the efficacy of the community genomics approach in serving as a public and environment health monitoring tool not only through the detection of through spatial variation in wastewater bacterial profile across the three sampling sites but also through the prediction of the pathogenic quantum in wastewater systems.
Acknowledgements
The authors acknowledge the Intramural grant of St. Xavier’s College (Autonomous) Kolkata awarded to Dr. Mahashweta Mitra Ghosh and DBT Grant (BT/INF/22/SP41296/2020) from the Department of Biotechnology, Government of India for the necessary infrastructure and other facilities. The authors are also thankful to Dr. Sayak Ganguli, Postgraduate and Research Department of Biotechnology, for his guidance in Metaanalysis.
Funding Sources
The authors acknowledge the Intramural grant of St. Xavier’s College (Autonomous) Kolkata awarded to Dr. Mahashweta Mitra Ghosh and DBT Grant (BT/INF/22/SP41296/2020) from the Department of Biotechnology, Government of India for the necessary infrastructure and other facilities.
Conflict of Interest
Authors declare that no conflict of interest exists.
Authors’ Contribution:
Rupsha Karmakar- Investigation, Data Collection, Writing- Original Draft, Data analysis
Kaustav Mondal- Formal data representation, Writing- Original Draft
Mahashweta Mitra Ghosh- Conceptualisation, Supervision, Writing- Review and Editing
Data Availability Statement
The manuscript incorporates three Wastewater Metagenomic datasets examined throughout this study. All were collected form Sequence Read Archive (SRA) available at NCBI database under the Accession Numbers- SRA: ERS946133, SRA: ERS613823 and SRA: SRS4735877.
Ethics Approval Statement
Not applicable.
References
- Sweileh W. M. Bibliometric analysis of peer-reviewed literature on climate change and human health with an emphasis on infectious diseases. Global Health. 2020;16(1):44. Published 2020 May 8. doi:10.1186/s12992-020-00576-1
CrossRef - Fouladkhah A. C, Thompson B, Camp J. S. The Threat of Antibiotic Resistance in Changing Climate. Microorganisms. 2020;8(5):748. Published 2020 May 16. doi:10.3390/microorganisms8050748
CrossRef - Rodríguez-González A, Zanin M, Menasalvas-Ruiz E. Public Health and Epidemiology Informatics: Can Artificial Intelligence Help Future Global Challenges? An Overview of Antimicrobial Resistance and Impact of Climate Change in Disease Epidemiology. Yearb Med Inform. 2019;28(1):224-231. doi:10.1055/s-0039-1677910
CrossRef - Larsson D. G. J, Flach C. F. Antibiotic resistance in the environment. Nat Rev Microbiol. 2022;20(5):257-269. doi:10.1038/s41579-021-00649-x
CrossRef - Liu X, Liu Z, Zhang Y, Jiang B. The Effects of Floods on the Incidence of Bacillary Dysentery in Baise (Guangxi Province, China) from 2004 to 2012. Int J Environ Res Public Health. 2017;14(2):179. Published 2017 Feb 12. doi:10.3390/ijerph14020179
CrossRef - IPCC. Special Report on Global Warming of 1.5°C (SR15). https://www.ipcc.ch/sr15/
- Berendes D, Knee J, Sumner T, et al. Gut carriage of antimicrobial resistance genes among young children in urban Maputo, Mozambique: Associations with enteric pathogen carriage and environmental risk factors. PLoS One. 2019;14(11):e0225464. Published 2019 Nov 22. doi:10.1371/journal.pone.0225464
CrossRef - MacFadden D. R, McGough S. F, Fisman D, Santillana M, Brownstein J. S. Antibiotic Resistance Increases with Local Temperature. Nat Clim Chang. 2018;8(6):510–514. https://doi.org/10.1038/s41558-018-0161-6
CrossRef - Burnham J. P. Climate change and antibiotic resistance: a deadly combination. Ther Adv Infect Dis. 2021;8:2049936121991374. Published 2021 Feb 15. doi:10.1177/2049936121991374
CrossRef - Mohsin M, Shad A. A, Ali J. Antimicrobial resistance, food systems and climate change. InSustainable Agriculture Reviews 46: Mitigation of Antimicrobial Resistance Vol 1 Tools and Targets 2020. p. 59-81. Cham: Springer International Publishing
CrossRef - Hughes J, Cowper-Heays K, Olesson E, Bell R, Stroombergen A. Impacts and implications of climate change on wastewater systems: A New Zealand perspective. Clim. Risk Manag. 2021;31:100262.
CrossRef - Bürgmann H, Frigon D, Gaze W, et al. Water and sanitation: an essential battlefront in the war on antimicrobial resistance. FEMS Microbiol Ecol. 2018;94(9):10.1093/femsec/fiy101. doi:10.1093/femsec/fiy101
CrossRef - Heljanko V, Tyni O, Johansson V, et al. Clinically relevant sequence types of carbapenemase-producing Escherichia coli and Klebsiella pneumoniae detected in Finnish wastewater in 2021-2022. Antimicrob Resist Infect Control. 2024;13(1):14. Published 2024 Jan 30. doi:10.1186/s13756-024-01370-z
CrossRef - Mavugara R, Matsa M. M. Resource recovery from municipal wastewater treatment plants: the Zimbabwean Perspective. Circular Economy and Sustainability. 2024;4(1):363-386.
CrossRef - Singh M, Karmakar R, Ganguli S, Ghosh M. M. Report of Antibiotic Resistance in Urban and Rural Wastewaters from West Bengal, India. Journal of Pharmaceutical Research International. 2021;33(53A):274–285.
CrossRef - Ondov B. D, Bergman N. H, Phillippy A. M. Interactive metagenomic visualization in a Web browser. BMC Bioinformatics. 2011;12:385. Published 2011 Sep 30. doi:10.1186/1471-2105-12-385
CrossRef - Oliveros J. C. Venny. An interactive tool for comparing lists with Venn's diagrams. 2007. https://bioinfogp.cnb.csic.es/tools/venny/index.html.
- Chong J, Liu P, Zhou G, Xia J. Using MicrobiomeAnalyst for comprehensive statistical, functional, and meta-analysis of microbiome data. Nat Protoc. 2020;15(3):799-821. doi:10.1038/s41596-019-0264-1
CrossRef - Nagpal S, Haque M. M, Singh R, Mande S. S. iVikodak-A Platform and Standard Workflow for Inferring, Analyzing, Comparing, and Visualizing the Functional Potential of Microbial Communities. Front Microbiol. 2019;9:3336. Published 2019 Jan 14. doi:10.3389/fmicb.2018.03336
CrossRef - Rossati A. Global Warming and Its Health Impact. Int J Occup Environ Med. 2017;8(1):7-20. doi:10.15171/ijoem.2017.963
CrossRef - Kaba H. E. J, Kuhlmann E, Scheithauer S. Thinking outside the box: Association of antimicrobial resistance with climate warming in Europe - A 30 country observational study. Int J Hyg Environ Health. 2020;223(1):151-158. doi:10.1016/j.ijheh.2019.09.008
CrossRef - Skariyachan S, Mahajanakatti A. B, Grandhi N. J, et al. Environmental monitoring of bacterial contamination and antibiotic resistance patterns of the fecal coliforms isolated from Cauvery River, a major drinking water source in Karnataka, India. Environ Monit Assess. 2015;187(5):279. doi:10.1007/s10661-015-4488-4
CrossRef - Cruz-Paredes C, Tájmel D, Rousk J. Can moisture affect temperature dependences of microbial growth and respiration? Soil Biol Biochem. 2021;156:108223.
CrossRef - Wu G. D, Chen J, Hoffmann C, et al. Linking long-term dietary patterns with gut microbial enterotypes. Science. 2011;334(6052):105-108. doi:10.1126/science.1208344
CrossRef - Africans, Diets of. Nutrition and Well-Being A to Z. . Retrieved July 28, 2022, from https://www.encyclopedia.com/food/news-wires-white-papers-and-books/africans-diets
- Kumar D, Patel Z, Pandit P, Pandit R, et al. Textile industry wastewaters from Jetpur, Gujarat, India, are dominated by Shewanellaceae, Bacteroidaceae, and Pseudomonadaceae harboring genes encoding catalytic enzymes for textile dye degradation. Front. environ. sci. 2021;9:720707.
CrossRef - Santoru M. L, Piras C, Murgia A, et al. Cross sectional evaluation of the gut-microbiome metabolome axis in an Italian cohort of IBD patients [published correction appears in Sci Rep. 2018 Mar 19;8(1):4993]. Sci Rep. 2017;7(1):9523. Published 2017 Aug 25. doi:10.1038/s41598-017-10034-5
CrossRef - Chen Y, Gui Q, Xu Q, Lv T, Gu S, Shen P, et al. Clostridium difficile Infection Among Hospitalized Chronic Hepatitis B Virus-Infected Patients in a Chinese Hospital. Jundishapur J. Microbiol. 2018;11(9).
CrossRef - Lee N. Y, Suk K. T. The Role of the Gut Microbiome in Liver Cirrhosis Treatment. Int J Mol Sci. 2020;22(1):199. Published 2020 Dec 28. doi:10.3390/ijms22010199
CrossRef - Su Z, Lu L, Chen F, Chen J, Chen X. Gut Microbiota and Sunitinib-Induced Diarrhea in Metastatic Renal Cell Carcinoma: A Pilot Study. Cancer Manag Res. 2021;13:8663-8672. Published 2021 Nov 20. doi:10.2147/CMAR.S328451
CrossRef - Ji D. D, Huang I. H, Lai C. C, et al. Prevalence and characterization of enterotoxigenic Bacteroides fragilis and toxigenic Clostridium difficile in a Taipei emergency department. J Microbiol Immunol Infect. 2017;50(1):83-89. doi:10.1016/j.jmii.2014.12.005
CrossRef - Climate of India. https://en.wikipedia.org/wiki/Climate_of_India
- UNDP. Zimbabwe. Climate Change Adaptation. https://www.adaptation-undp.org/explore/eastern-africa/zimbabwe
- Bhavnani D, Goldstick J. E, Cevallos W, Trueba G, Eisenberg JN. Impact of rainfall on diarrheal disease risk associated with unimproved water and sanitation. Am J Trop Med Hyg. 2014;90(4):705-711. doi:10.4269/ajtmh.13-0371
CrossRef - Mills R. H, Dulai P. S, Vázquez-Baeza Y, et al. Multi-omics analyses of the ulcerative colitis gut microbiome link Bacteroides vulgatus proteases with disease severity. Nat Microbiol. 2022;7(2):262-276. doi:10.1038/s41564-021-01050-3
CrossRef - Aamodt G, Bengtson M. B, Vatn M. H. Can temperature explain the latitudinal gradient of ulcerative colitis? Cohort of Norway. BMC Public Health. 2013;13:530. Published 2013 May 31. doi:10.1186/1471-2458-13-530
CrossRef - Hricová K, Röderová M, Fry?ák P, et al. Prevalence of Vancomycin-Resistant Enterococci and Antimicrobial Residues in Wastewater and Surface Water. Life (Basel). 2021;11(12):1403. Published 2021 Dec 15. doi:10.3390/life11121403
CrossRef - Tarrant C, Colman A. M, Jenkins D. R, et al. Drivers of Broad-Spectrum Antibiotic Overuse across Diverse Hospital Contexts-A Qualitative Study of Prescribers in the UK, Sri Lanka and South Africa. Antibiotics (Basel). 2021;10(1):94. Published 2021 Jan 19. doi:10.3390/antibiotics10010094
CrossRef







